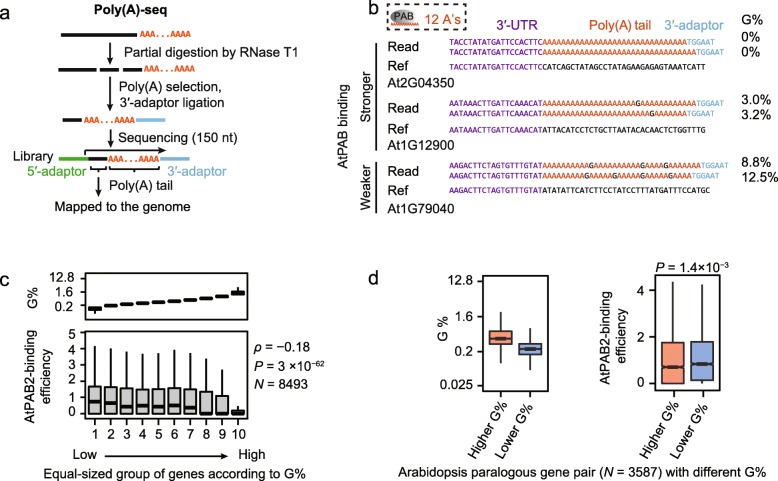
Fig. 3.
G-content contributes to the binding variance of AtPABs. a Schematic of the poly(A)-seq experiment that directly sequences the poly(A) tails of mRNAs. High-quality reads with recognizable Illumina 3′-adaptor sequences were used for further analysis. b Example reads from the poly(A)-seq show different propensities for AtPAB binding among genes. Yeast Pab1p and human PABP require at least 12 and 11 consecutive A’s for binding, respectively. Guanosines can inhibit AtPAB binding by cutting the poly(A) tail into fragments with < 12 consecutive A’s. Ref represents the reference Arabidopsis genome. c The G% was negatively correlated with the AtPAB-binding efficiency of a gene. Genes were divided into ten equal-size groups according to the average G% in the poly(A) tails. P values were given by the Spearman’s correlation. d Paralogous genes with lower G% in the poly(A) tail exhibited higher AtPAB2-binding efficiencies than their within-species paralogs with higher G% in the poly(A) tails. P values were given by the paired Mann-Whitney U test