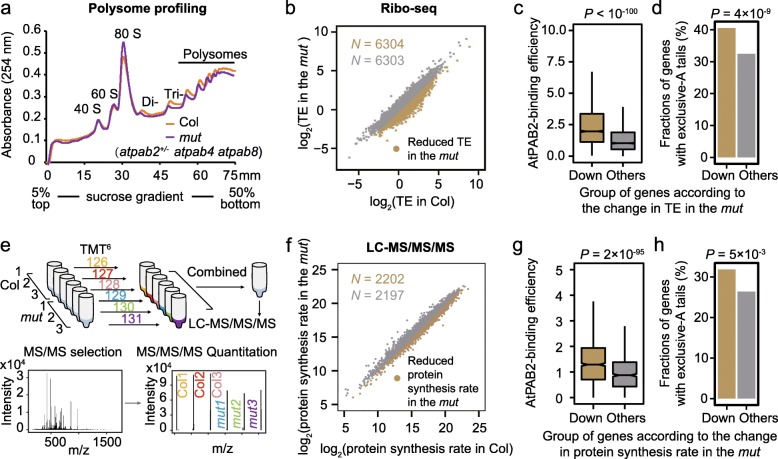
Fig. 5.
AtPABs enhance translational efficiency. a Polysome profiling shows the global reduction in TE in the mut. The x-axis indicates the detecting distance from 0 to 75 mm of the 5–50% sucrose gradient. b, c Genes showing reduced TE in the mut (b) exhibited significantly higher AtPAB2-binding efficiencies (c). The TE was calculated with the ribo-seq data. Genes with decreased TE in the mut (brown dots) were defined as those with TE fold change smaller than the median. P value in c was given by the Mann-Whitney U test. d Genes with exclusive adenosines in the poly(A) tail tended to be present in the gene group showing the downregulation of TE in the mut (defined in b). e Schematic of the TMT (Tandem Mass Tag)-based quantitative proteomics analysis. Protein samples extracted from three biological replicates of Col and the mut were labeled with TMT 6 plex reagent separately. f–h The protein synthesis rate was calculated as the protein level of a gene normalized to its mRNA level, and similar results were obtained as those in b–d