Abstract
Background
Biosynthetic gene clusters produce a wide range of metabolites with activities that are of interest to the pharmaceutical industry. Specific interest is shown towards those metabolites that exhibit antimicrobial activities against multidrug-resistant bacteria that have become a global health threat. Genera of the phylum Firmicutes are frequently identified as sources of such metabolites, but the biosynthetic potential of its Virgibacillus genus is not known. Here, we used comparative genomic analysis to determine whether Virgibacillus strains isolated from the Red Sea mangrove mud in Rabigh Harbor Lagoon, Saudi Arabia, may be an attractive source of such novel antimicrobial agents.
Results
A comparative genomics analysis based on Virgibacillus dokdonensis Bac330, Virgibacillus sp. Bac332 and Virgibacillus halodenitrificans Bac324 (isolated from the Red Sea) and six other previously reported Virgibacillus strains was performed. Orthology analysis was used to determine the core genomes as well as the accessory genome of the nine Virgibacillus strains. The analysis shows that the Red Sea strain Virgibacillus sp. Bac332 has the highest number of unique genes and genomic islands compared to other genomes included in this study. Focusing on biosynthetic gene clusters, we show how marine isolates, including those from the Red Sea, are more enriched with nonribosomal peptides compared to the other Virgibacillus species. We also found that most nonribosomal peptide synthases identified in the Virgibacillus strains are part of genomic regions that are potentially horizontally transferred.
Conclusions
The Red Sea Virgibacillus strains have a large number of biosynthetic genes in clusters that are not assigned to known products, indicating significant potential for the discovery of novel bioactive compounds. Also, having more modular synthetase units suggests that these strains are good candidates for experimental characterization of previously identified bioactive compounds as well. Future efforts will be directed towards establishing the properties of the potentially novel compounds encoded by the Red Sea specific trans-AT PKS/NRPS cluster and the type III PKS/NRPS cluster.
Electronic supplementary material
The online version of this article (10.1186/s12864-019-6065-7) contains supplementary material, which is available to authorized users.
Keywords: Virgibacillus, Antimicrobial, Biosynthetic gene clusters, Genome-mining, Nonribosomal peptides, Polyketides, Bacteriocins, Lanthipeptides, Bioinformatics
Background
Biosynthetic gene clusters (BGCs), made up of multi-enzymatic, multi-domain megasynthases, are often of interest in genome-mining. For instance, the most well studied modular clusters, nonribosomal peptide synthetases (NRPSs) and Polyketide Synthases (PKSs) are often associated with the synthesis of antitumor, antimicrobial, antifungal, and immunosuppressive products [1, 2]. Another BGC class is the ribosomally synthesized and posttranslationally modified peptides (RiPPs) [3] which include the extensively studied bacteriocins and lanthipeptides [3], both of which have known products with a wide spectrum of antimicrobial activity. In fact, lanthipeptides were initially termed lantibiotics because the first identified clusters exhibited antibiotic activity. But, as more and more clusters without antibiotic activity were discovered, the more generic term lanthipeptide came into use. A well-described example of anon-antibiotic lanthipeptide is SapB identified in Streptomyces coelicolor [4]. Nonetheless, other RiPPs also exhibit antimicrobial activity including thiopeptides, bottromycins, lipolanthines, etc.
Because of this knowledge and the increase in available sequenced genomes, methods have been developed to allow mining of sequenced genomes for these BGCs [5–11]. However, there are no studies that provide insights into genomic features in strains belonging to the Virgibacillus genus of the Firmicutes phylum. This is surprising owing to the fact that: 1/ the rod-shaped, endospore-forming species in this genus are both gram-stain variable and gram-stain positive, 2/ these strains exhibit an ability to adapt to diverse environments such as marine sediment [12], faeces [13] and fermented food [14, 15], 3/ these strains exhibit enzymatic and antimicrobial potential of interest to industry [15–20], and 4/ there is a large number of sequenced Virgibacillus genomes (31 at the time of conducting our study, with six complete genomes assembled).
We previously reported [20] that V. dokdonensis Bac330 and Virgibacillus sp. Bac332 exhibited antimicrobial activity against both Staphylococcus aureus (ATCC25923) and Pseudomonas syringae (dc3000), respectively, while V. halodenitrificans Bac324 displayed no such activity. These strains were isolated from Red Sea locations which are shown through metagenomic analysis [21] to harbor a rich repertoire of NRPS and PKS sequences. The nature of the environment (high salinity and temperature) can be a contributing factor to the horizontal transfer of mobile genetic elements contributing to strengthening the biosynthetic potential of its microbiome. Here, we identify features in these three Virgibacillus strains isolated from the Red Sea, which provide insights into strains’ biosynthetic potential encoded by their genomes. Based on the comparison with other strains from the same genus, we show specific genetic characteristics unique to the marine Virgibacillus strains, including Red Sea strains.
Results and discussion
Features of the genomes of the Red Sea Virgibacillus strains
Raw sequences of the three Virgibacillus genomes- V. dokdonensis Bac330 (CP033048), Virgibacillus sp. Bac332 (CP033046-CP033047), and V. halodenitrificans Bac324 (CP033049- CP033050)- showed that on average there are 133,749.6667 reads per genome (141,676, 148,002, 111,571 reads for V. dokdonensis Bac330, Virgibacillus sp. Bac332, and V. halodenitrificans Bac324, respectively). The read mean length was 7952 bp (253x genome coverage) for V. dokdonensis Bac330, 8939 bp (290x genome coverage) for Virgibacillus sp. Bac332 and 11,324 bp (311x genome coverage) for V. halodenitrificans Bac324. Assemblies of the reads showed that two of the strains have plasmids (Virgibacillus sp. Bac332 and V. halodenitrificans Bac324), while V. dokdonensis Bac330 only has one circular chromosome without plasmid. V. dokdonensis Bac330’s circular chromosome was 4,456,326 bp in length with 4163 predicted open reading frames (ORFs) (52.3% on the forward strand, and 47.7% on the reverse complement strand). V. halodenitrificans Bac324 circular chromosome was 4,063,118 bp in length with 4284 predicted ORFs (46.9% on the forward strand and 53.1% on the reverse complement one) and its plasmid was 312,876 bp in length. Virgibacillus sp. Bac332 circular chromosome was 4,561,556 bp in length with 4424 predicted ORFs (46.1% on the forward strand and 53.9% on the reverse complement one) and its plasmid was 65,691 bp in length. The total number of tRNAs was almost the same in the three genomes (63,64,64 tRNAs), while the number of rRNAs was 18 for both Virgibacillus sp. Bac332 and V. dokdonensis Bac330 but was 24 for V. halodenitrificans Bac324 (Table 1, Fig. 1).
Table 1.
Summary of the genomes and annotation of nine Virgibacillus strains
| Strain | Isolation site | Number of ORFs | Genome size (Mb) | GC% | Number of scaffolds | Genomic Islands % | # of rRNAs | # of tRNAs | # of unique genes |
|---|---|---|---|---|---|---|---|---|---|
| Virgibacillus sp. SK37 (Genbank accession number: CP007161) | Fish sauce mash | 3795 | 3.84 | 37.59 | 4 | 4.9 | 21 | 53 | 504 |
| Virgibacillus halodenitrificans PDB-F2 (Genbank accession number: CP017962) | Solid waste landfill | 3820 | 3.92 | 37.43 | 2 | 2.8 | 24 | 62 | 255 |
| Virgibacillus phasianinus LM2416 (Genbank accession number: CP022315) | Host Lophura swinhoei | 3966 | 4.07 | 39.50 | 1 | 4.1 | 21 | 64 | 648 |
| Virgibacillus necropolis LMG 19488 (Genbank accession number: CP022437) | Mural paintings | 4135 | 4.34 | 37.30 | 1 | 2.2 | 22 | 62 | 761 |
| Virgibacillus dok0064onensis 21D (Genbank accession number: CP018622) | Marine | 3939 | 4.26 | 36.60 | 1 | 5.4 | 18 | 63 | 435 |
| Virgibacillus sp. 6R (Genbank accession number: CP017762) | Coal bed | 4313 | 4.75 | 37.30 | 1 | 5.9 | 18 | 65 | 927 |
| Virgibacillus halodenitrificans Bac324 (Genbank accession number: CP033049) | Mangrove mud/Rabigh Harbour Lagoon | 4284 | 4.06 | 37.2 | 2 | 5.19 | 24 | 62 | 665 |
| Virgibacillus sp. Bac332 (Genbank accession number: CP033046) | mangrove mud | 4424 | 4.56 | 36.7 | 2 | 9.32 | 18 | 63 | 958 |
| Virgibacillus dokdonensis Bac330 (Genbank accession number: CP033048) | Mangrove mud | 4163 | 4.46 | 36.8 | 1 | 6.9 | 18 | 64 | 619 |
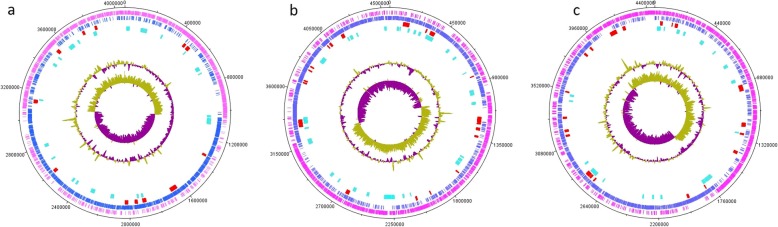
Fig. 1.
Circular plots of (a) Bac330 and (b) Bac324 and (c) Bac332 genomes, showing the overlap of biosynthetic genes and genomic islands. The tracks show the following features starting from the outermost track; 1st track (pink): genes on the positive strand; 2nd track (blue): genes on the negative strand; 3rd track (yellow): biosynthetic gene clusters; 4th track (red): horizontally transferred genes; 5th track (cyan): genes in prophage regions; 6th track: GC-plot where purple and green correspond to below and above average GC content, respectively; 7th track: GC-skew where purple and green correspond to below and above average GC-skew, respectively
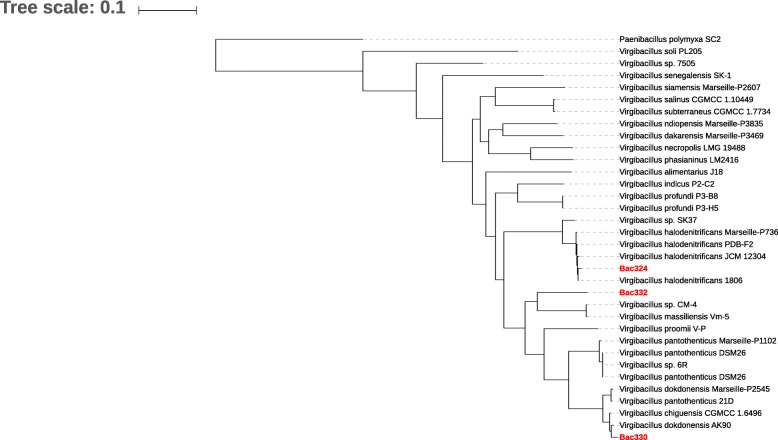
For phylogenetic placement of the three Red Sea strains, a phylogenetic tree was generated using 606 single-copy genes (Fig. 2). We included a total of 31 complete and incomplete Virgibacillus genomes for a higher resolution in the placement. We also used Paenibacillus polymyxa as the outgroup.
Fig. 2.
Maximum-likelihood phylogenetic tree of 32 genomes constructed using 606 single-copy genes. Paenibacillus polymyxa was used as the outgroup. The tree shows the phylogenetic proximity of the Red Sea Virgibacillus strains in the Virgibacillus group
The resulting tree (Fig. 2) shows the phylogenetic proximity Bac330, Bac324, and Bac332 to Virgibacillus strains as indicated by the 16S rRNA analysis reported in a previous study [20].
To identify the unique functional groups in the three complete Virgibacillus genomes from the Red Sea, we investigated the accessory genome of these isolates and searched for orthologous families that have genes exclusively present in any genome of the three Red Sea Virgibacillus strains and are consistently absent from the six complete publicly available Virgibacillus genomes. The analysis showed that amongst the 11,135 gene families that constitute the pan-genome of the analyzed strains, there are 958 unique genes in Virgibacillus sp. Bac332. There are 665 unique genes in V. halodenitrificans Bac324 and 619 in V. dokdonensis Bac330 (See Table 1). The number of unique genes in the non-Red Sea Virgibacillus strains is 255 in V. halodenitrificans PDB-F2, 761 in V. necropolis LMG 19488, 435 in V. dokdonensis 21D, 504 in Virgibacillus sp. sk37, 927 in Virgibacillus sp. 6R and 648 in V. phasianinus LM2416. Thus, Virgibacillus sp. Bac332 has the highest number of unique genes among the nine genomes.
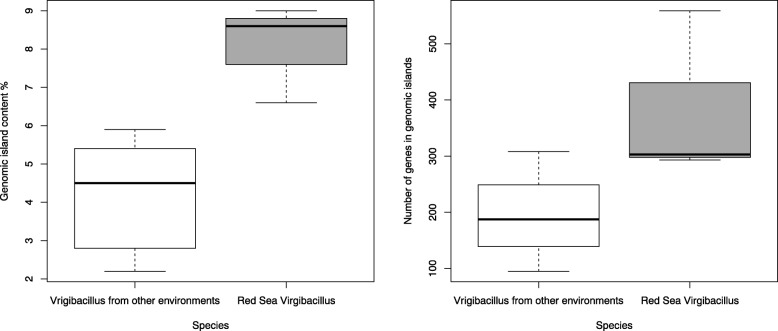
Since horizontal gene transfer is considered central to microbes’ ability to adapt to an ecological niche, we also predicted the genomic islands (GIs) in the analyzed genomes to increase our understanding as to how the three Virgibacillus strains have potentially acquired genomic elements adding to their biosynthetic capacity. Interestingly, the chromosomal sequence of Virgibacillus sp. Bac332 has the highest number of GIs with a total of 434,257 bp of DNA (21 GIs harboring 559 genes), equivalent to 9% of the genome size. GI prediction also identified 210,858 bp of the chromosomal sequence in the genome of V. halodenitrificans Bac324 (13 GIs harboring 293 genes) and 309,859 bp in V. dokdonensis Bac330 (19 GIs harboring 303 genes) amounting to 5.19 and 6.9% of the genomes, respectively. On average, there is 178,642 bp of DNA sequence in GI predicted regions in any of the publicly available chromosomal sequences of the six non-Red Sea Virgibacillus strains, where V. necropolis LMG 19488 has the lowest number of GIs (6 GIs) extending over 96,572 bp of DNA and harboring 95 genes. On the other hand, the genomes of the Red Sea isolates have on average 318,325 bp of GI sequence per genome (Fig. 3).
Fig. 3.
Boxplot of genomic island content and number of genes in genomic islands falling in the public Virgibacillus strains (white) and Red Sea Virgibacillus (grey) using both the size of the DNA regions in which predicted GIs fall as well as the number of predicted genes
These analyses show that despite the large size of the core genome (1654 ORFs accounting for 40% of the average number of ORFs), one Red Sea strain has a large number of unique genes that are not orthologous to any other gene in the studied Virgibacillus genomes, at least using the imposed identity threshold (see the Materials and Methods). Moreover, the strikingly high GI content in the genomes of Virgibacillus sp. Bac332 and V. dokdonensis Bac330, along with the high number of biosynthetic genes and unique genes, collectively hint toward the presence of unique functional genomic features in marine Virgibacillus strains compared to other genomes used in this study. Although we could not collectively discriminate strains based on environment type only, we have shown increased uniqueness in the genomic features of one of the Red Sea strains, Virgibacillus sp. Bac332.
Exploring the biosynthetic potential of the Red Sea Virgibacillus strains
To identify unique biosynthetic elements in the Red Sea strains that might indicate specific functions as a result of environmental adaptation, we had to confirm that the biosynthetic features identified in marine isolates, including ones from the Red Sea, are not present in closely related species. To do so, we included the six complete Virgibacillus species along with the three Red Sea strains for the evaluation (Table 1).
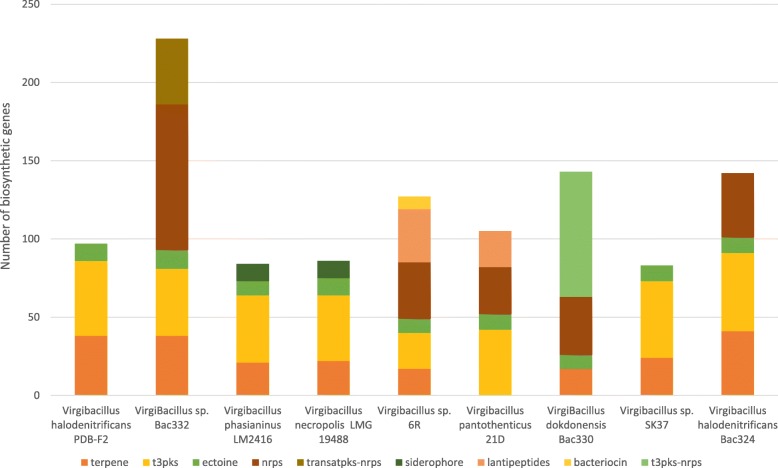
On average, each of the analyzed genomes comprises 24 putative biosynthetic gene clusters (BGCs) that were predicted by antiSMASH or ClusterFinder. The clusters predicted to fall in known classes of BGCs are mainly encoding for proteins related to the following biosynthetic classes: terpene, type III PKS, ectoine, NRPS, trans-acyltransferase PKS/NRPS, siderophore, lanthipeptide, bacteriocin, and type III PKS/NRPS (Fig. 4). Of interest to this study are two classes of secondary metabolites known for their use in different applications of pharmaceutical and industrial interest: 1/ modular clusters encompassing mainly NRPSs and modular PKS, and 2/ ribosomally synthesized peptides, namely bacteriocins and lanthipeptides.
Fig. 4.
Distribution of genes in biosynthetic gene clusters in nine Virgibacillus genomes. Strains are color-coded as per the legend. The distribution clearly shows that genomes with the highest number of genes in BGCs are in the Virgibacillus Red Sea isolates
Gene cluster families in Virgibacillus strains
A total of 215 BGCs (173 putative clusters and 42 in known BGC classes) with 3631 genes were classified into 35 groups (also referred to as gene cluster families GCFs) using scoring similarity networks as implemented in BiG-SCAPE [22] .
Interestingly, only three gene cluster families were assigned to clusters that produce known products or have a similar pathway using threshold similarity of 60% (these include paeninodin, locillomycin, and ectoine). Most notably, there is at least one terpene, type III PKS, and ectoine cluster in all of the genomes. Only one bacteriocin encoding BGC is identified in Virgibacillus sp. 6R, while lipopeptides were only identified in Virgibacillus sp. 6R and V. dokdonensis 21D. Siderophores were identified in V. phasianinus LM2416 and V. necropolis LMG 19488 only. The diversity of the distribution of BGC types across the Virgibacillus genomes, in spite of phylogenetic proximity, is an indication of the acquisition of genomic elements that enable biosynthetic routes of various products at different isolation sites. To investigate the emergence of environment-specific clusters, we looked at gene cluster families that are not shared by strains of the same species but shared between strains from different phylogenetic groups in the tree. In total, out of 35 gene cluster families, 10 are shared between at least one of the three Red Sea isolates and V. dokdonensis strain 21D; the only other marine isolate in the analysis (Additional file 1: Figure S1). Specifically, 6 of these clusters are exclusively shared between V. dokdonensis strain 21D and V. dokdonensis Bac330 or Virgibacillus sp. Bac332. Interestingly, other strains that have repeated patterns of shared clusters are either strains of the same species or strains falling in the same phylogenetic group. Additionally, Bac332 shares one cluster with V. halodenitrificans Bac324 and V. halodenitrificans PDB-F2, despite falling in different groups. Unfortunately, since none of these clusters have assigned products, we could not elaborate on the putative functions of these clusters.
Modular clusters and ribosomally synthesized and posttranslationally modified peptides (RiPPs)
In order to identify any exclusive modular clusters in the genomes of the marine isolates, we categorized the clusters based on gene homology and product type and assessed their distributions in the genomes. We found that out of a total of eight NRPSs and modular PKSs collectively identified in the nine Virgibacillus genomes, six are in the Red Sea Virgibacillus strains (two in V. dokdonensis Bac330, three in Virgibacillus sp. Bac332 and one in V. halodenitrificans Bac324) (Additional file 2: Table S1). The other two modular clusters are in Virgibacillus sp. 6R and in V. dokdonensis 21D. Only one of the clusters maps to known BGCs using a 60% similarity threshold (a hybrid trans-AT PKS/NRPS cluster in Bac332 with 64% similarity to Locillomycin). One NRPS cluster shares a number of structural features in four strains (Bac330, Bac324, 6R, and 21D), albeit with some variations hindering their assignment as the same cluster. Specifically, Bac324 and 6R have clusters in the same GCF with the highest similarity, while the cluster in 6R is the most variant with a single KS domain, and a cis-acting AT domain. The clusters in Bac324 and 6R have a combination of shared condensation (c) domains, adenylation (A) domains, as well as acylotransferase (AT) domains (that are putatively acting in trans due to their organization in the cluster as standalone domains). The presence of PKS domains such as ketosynthase (KS), ketoreductase (KR) in Bac330, 6R and 21D indicates that these could be a hybrid PKS/NRPS clusters. It is noteworthy that none of the hybrid PKS/NRPS clusters or NRPS ones in Virgibacillus sp. Bac332 share the same modular structure as other modular clusters in other Virgibacillus strains.
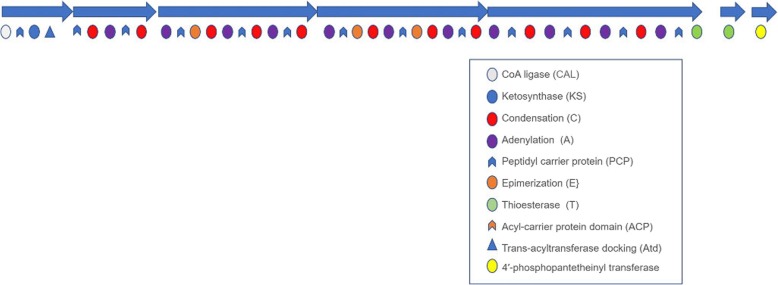
One uniquely structured cluster that has both PKS and NRPS domains is identified in the Red Sea Virgibacillus sp. Bac332. The cluster is characterized by the presence of four NRPS synthases composed of Adenylation (A), condensation (C) Epimerization (E), peptidyl carrier protein (PCP) domains and a thioesterase (T) domain. It also has one modular PKS synthase with a ketosynthase (KS) domain, a CoA ligase (CAL) domain and a trans-AT binding site (Fig. 5). There are also two single-domain peptides in the cluster: one with a 4′-phosphopantetheinyl transferase and one with another thioesterase. The cluster has a 64% similarity to the BGC encoding locillomycin. This product was previously reported in Bacillus subtilis 916 to be encoded by an unusual synthase that does not follow the collinearity rule of assembly-line clusters [23]. Despite the high similarity, we noted that the cluster in Virgibacillus sp. Bac332 has five additional NRPS modules, making it larger than the one in B. subtilis 916 (86 Kb in Virgibacillus sp. Bac332 and 38 Kb in Bacillus subtilis 916). We also noted that out of the five mega-synthases that make up the trans-AT PKS/NRPS cluster, four mapped to all of the known modular proteins in the locillomycin BGC (LocA, LocB, LocC, and LocD). The fact that the fifth 12,990 bp mega-synthase does not have an ortholog in the locillomycin cluster indicates putative novelty of the synthesized product.
Fig. 5.
Structure of the hybrid PKS/NRPS cluster present in Bac332. Biosynthetic genes are identified with blue arrows. Domains abbreviations and color codes are shown in the legend
There is also a large, uniquely-structured, hybrid Type III PKS/NRPS cluster in V. dokdonensis Bac330, and Virgibacillus dokdonensis 21D. The cluster is composed of 81 genes extending over 91 Kb of DNA. Specifically, there are three modular NRPS genes and three single-domain proteins as part of the type III PKS component of the cluster. This component has a 14% similarity to the cereulide BGC. However, none of the genes overlapping to the cereulide cluster are modular core NRPS ones (the genes mapping to the cereulide cluster are genes with an alpha/beta hydrolase domain and a gene encoding an ABC-2 type transport system binding protein). There are two bacteriocin and two class II lanthipeptide clusters in the analyzed genomes but none were identified in the Red Sea Virgibacillus species). The bacteriocin cluster has a glutamine synthetase, a merR family transcriptional regulator glutamine synthetase repressor, and one bacteriocin biosynthetic gene.
Taken together, when estimating the biosynthetic potential of these strains, we find that only a few of the identified clusters in the analyzed strains could be assigned to known products despite a total of 215 BGCs identified in the considered Virgibacillus genomes. Specifically, ~ 79% of the clusters per genome are putative and do not fall in known classes of BGCs. This is an indication that the biosynthetic potential of Virgibacillus species should be explored in much more details; there is an increasing number of publicly available genomes. Nonetheless, our results suggest that Virgibacillus sp. Bac332 has the highest number of modular genes falling in two NRPS clusters and a hybrid trans-AT PKS/NRPS cluster. Moreover, no bacteriocins or lanthipeptides were identified in the Red Sea isolates. The prevalence of modular clusters and low frequency of RiPPs in the Red Sea isolates could be part of an array of phenotypes required to adapt to the specific ecological conditions of the Red Sea. Moreover, the specific locations from which these strains were collected are in the Rabigh Harbor Lagoon, a substantial part of which has been converted into a harbor serving the Petro Rabigh petrochemical and refining complex and is therefore expected to be contaminated.
We specifically utilized the antiSMASH ClusterBlast module, as it allows us to assess the homology of the genes in the cluster to all genomes deposited in NCBI, either complete or draft. In Additional file 2: Table S1, we report the species with the top hits for these clusters. The fact that only a few of the top hits for the clusters in the Red Sea strains are Virgibacillus species supports our conclusions about the potential uniqueness of these clusters within the Virgibacillus genus. That is, most of the NRPS clusters (four out of six clusters) in the Red Sea strains have no match to any of the Virgibacillus genomes (complete or draft), but rather match some other distant Bacillus genomes, most of which are from saline environments. Moreover, one of the clusters in BAC332 had only 7% percent of its genes similar to a cluster in Bacillus nakamurai strain NRRL B-41091, indicating indeed that this is a unique cluster. Other clusters in the Red Sea strain have more than 60% identity to clusters in Bacillus megaterium MSP20.1 and Tumebacillus sp. AR23208. Modular clusters from the remaining Virgibacillus genomes (6R and 21D) were similar to clusters in Marininema mesophilum strain DSM 45610 and Virgibacillus dokdonensis strain Marseille-P2545, respectively. It is fair to summarize from the above analysis that at least one cluster in Bac332, is acquired by the strain as part of an environmental adaptation mechanism. This conclusion does not negate or conflict with the fact that other marine species from other environments might acquire similarly-structured clusters.
To further support the notion that Red Sea specific genes may be truly unique, we further performed homology analysis of modular genes (i.e., genes with NRPS or PKS domains) in the 6 clusters against MarCat from the MAR databases [24] (with Evalue:<1e-5; percent identity> 35%; and bitscore > 50, see Additional file 3: Table S2). Almost all of the modular genes have a hit greater than 30%, albeit one cluster often has different hits in different metagenome samples. This cannot be interpreted as an incomplete cluster as we can potentially attribute it to the incomplete nature of sequenced metagenomes. However, since overall, there is at least one significant hit in one metagenome per cluster, it is fair to attribute the uniqueness of the biosynthetic genes in the Red Sea genomes to marine environments in general.
Biosynthetic gene clusters in genomic islands
The majority of the NRPS clusters in the Red Sea Virgibacillus genomes were found to fall in predicted GIs. Specifically, the NRPS identified in Bac332 is falling in a GI extending from 3,307,119 to 3,365,396 bp; while the NRPS cluster identified in Bac324 falls within the plasmid sequence identified in that genome. The NRPS cluster in V. dokdonensis Bac330 was found to overlap with a GI extending from 2,774,299 to 2,829,247 bp.
The fact that the majority of the identified NRPS clusters in the Red Sea isolates are part of GI regions motivated further investigation of the GIs in other Virgibacillus strains that have NRPS genes. Interestingly, we identified that the NRPS clusters in Virgibacillus sp. 6R (isolated from a Coal Bed) and Virgibacillus pantothenticus 21D (isolated from a marine environment) indeed are part of GIs that cover the region extending from1,078,422 to 1,129,3882 bp and from 2,579,235 to 2,619,409 bp, respectively. It is also noted that in most of the BGCs, there seems to be an overlap between the regions surrounding predicted BGC and GIs, which might indicate the acquisition of specialized genes necessary for the cluster, most of which are either transport elements or uncharacterized genes.
Conclusions
Despite the availability of a total of 30 publicly available Virgibacillus genomes (at the time of the study), the biosynthetic potential of these strains has not been sufficiently explored. Species from marine environments, such as the Red Sea [25], were shown to harbor a number of promising clusters for the biosynthesis of modular antimicrobial clusters with unique structural properties. Here, we are analyzing Virgibacillus strains isolated from the Red Sea, along with publicly available Virgibacillus genomes, for their biosynthetic capabilities, leveraging the availability of complete genomes of the Red Sea isolates. To do so, we first computed the core and pangenome of nine Virgibacillus strains including the Red Sea ones. Our analysis shows that most of modular NRPS clusters in the analyzed genomes of Virgibacillus strains are part of horizontally transferred genomic regions in genomic islands. We also show that two of the Red Sea isolates (V. dokdonensis Bac330 and Virgibacillus sp. Bac332) collectively have more modular genes compared to all the analyzed genomes, indicating the possibility of the emergence of specific biosynthetic genomic elements in the genomes of these isolates in response to environmental selection in unique marine environments. Future efforts will be directed towards unveiling the biotechnological advantages of these isolates and in this process establishing the properties of the compounds encoded by the Red Sea specific trans-AT PKS/NRPS cluster and other NRPS clusters in Bac332. This would serve to highlight the novelty of the bioactive compounds, in terms of both function and structure, compared to other known compounds.
Methods
DNA extraction, sequencing, assembly, and annotation
A detailed description of sampling, isolation, and purification of the strains is available in [20]. Strains Bac330, Bac324 and Bac332 were isolated from mangrove mud samples collected from the Rabigh Harbor Lagoon in Saudi Arabia (39°0′35.762′′E, 22°45′5.582′′ N). Genome sequencing was performed at the Core Laboratory sequencing facility at KAUST using the PacBio RS II sequencing platform (Pacific Biosciences, USA) and assembled using PacBio’s SMRT Analysis pipeline v2.3.0. using default parameters. Prodigal [26] was used as the gene prediction method and genome annotation was completed using the Automatic Annotation of Microbial Genomes pipeline (AAMG) [27] with default parameters. A detailed description of sequencing, assembly, and annotation are available in [25].
Comparative analysis of genomic features, genomic islands and biosynthetic genes
The core and pan-genomes were computed using GET_HOMOLOGUES v1.3 with the MCL option (−t 0 for core genome and -t all for pan-genome). We used a similarity threshold of 70%, alignment length coverage of 75% and e value of 1e-6. The number of core, shared and unique genes were visualized in an Upset fig. [28] as implemented in Intervene [29]. Prediction of GIs was completed using IslandViewer v4 [30, 31]. Circular visualization of the genomes and annotated features were plotted using DNAPlotter [32]. The phylogenetic tree was constructed using OrthoFinder v2.2.1 [33] utilizing gene trees for each orthogroup, with default settings, and visualized in iTOL [34]. Biosynthetic gene clusters were predicted using antiSMASH v3.0 [35] and cluster genes were mapped back to the MIBiG database in order to assign products to known clusters, with manual inspection of the focality of these alignments to core biosynthetic genes, as opposed to other accessory ones. Additionally, the ClusterFinder algorithm was also used to identify putatively novel clusters [36], Big-SCAPE [22] was used to group the cluster into families and Cytoscape v3.6.1 was used to visualize the resultant network of cluster families.
Additional files
Figure S1. Network visualization of 35 gene cluster families in Virgibacillus, showing that few groups are found in the majority of Virgibacillus genomes and none are found in all nine genomes (DOCX 100 kb)
Table S1. Features of modular clusters identified in Virgibacillus genomes (DOCX 16 kb)
Table S2. Homology analysis of the modular genes in six clusters identified in Red Sea Virgibacillus strains to marine metagenomes. (DOCX 13 kb)
Acknowledgments
The authors wish to acknowledge the experimental support from the King Abdullah University of Science and Technology (KAUST) Bioscience Core Laboratory. We would also like to thank L’Oréal - UNESCO for awarding the first author (GO) the ‘For Women in Science fellowship’, as a recognition, partly, for this work.
Abbreviations
- A
Adenylation
- AAMG
Automatic annotation of microbial genomes pipeline
- ACP
Acyl carrier protein
- AT
Acyltransferase
- BGC
Biosynthetic gene clusters
- C
Condensation
- CAL
CoA ligase
- E
Epimerization
- GCF
Gene cluster family
- GIs
Genomic islands
- KR
Ketoreductase
- KS
Ketosynthase
- ML
Maximum likelihood
- NRPS
Nonribosomal peptide synthetase
- PCP
Peptidyl carrier protein
- PKS
Polyketide synthase
- RiPP
Ribosomally synthesized and post-translationally modified peptide
- SMRT
Single-molecule real-time
- T
Thioesterase
Authors’ contributions
The study was conceived and designed by VBB, GO, and ME. Data was generated by AB, HH, and FFL. Data analysis was performed by GO, SB, IM, and ME. The manuscript was written by GO, IM, TG, VBB, and ME. All authors read and approved the final version of the manuscript.
Funding
The research reported in this publication was supported by King Abdullah University of Science and Technology (KAUST) through the Awards Nos. FCC/1/1976-02-01, FCC/1/1976-17-01, BAS/1/1606-01-01, FCC/1/1976-03-01, FCC/1/1976-20-01, FCC/1/1976-16-01 and BAS/1/1062-01-01 from the Office of Sponsored Research (OSR). The funding bodies did not participate in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Availability of data and materials
All data used in this study have been included in this article.
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
GO, SB, RR, AB, FFL, TG, HH, IM, VBB, and ME declare no competing interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Ghofran Othoum, Email: ghofran.othoum@kaust.edu.sa.
Salim Bougouffa, Email: salim.bougouffa@kaust.edu.sa.
Ameerah Bokhari, Email: ameerah.bokhari@kaust.edu.sa.
Feras F. Lafi, Email: Feras.Lafi@zu.ac.ae
Takashi Gojobori, Email: takashi.gojobori@kaust.edu.sa.
Heribert Hirt, Email: heribert.hirt@kaust.edu.sa.
Ivan Mijakovic, Email: ivan.mijakovic@chalmers.se.
Vladimir B. Bajic, Email: vladimir.bajic@kaust.edu.sa
Magbubah Essack, Phone: +966 (54) 470 0746, Email: magbubah.essack@kaust.edu.sa.
References
- 1.Walsh CT. Polyketide and nonribosomal peptide antibiotics: modularity and versatility. Science. 2004;303(5665):1805–1810. doi: 10.1126/science.1094318. [DOI] [PubMed] [Google Scholar]
- 2.Felnagle EA, Jackson EE, Chan YA, Podevels AM, Berti AD, McMahon MD, Thomas MG. Nonribosomal peptide synthetases involved in the production of medically relevant natural products. Mol Pharm. 2008;5(2):191–211. doi: 10.1021/mp700137g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arnison PG, Bibb MJ, Bierbaum G, Bowers AA, Bugni TS, Bulaj G, Camarero JA, Campopiano DJ, Challis GL, Clardy J, et al. Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat Prod Rep. 2013;30(1):108–160. doi: 10.1039/C2NP20085F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kodani S, Hudson ME, Durrant MC, Buttner MJ, Nodwell JR, Willey JM. The SapB morphogen is a lantibiotic-like peptide derived from the product of the developmental gene ramS in Streptomyces coelicolor. Proc Natl Acad Sci U S A. 2004;101(31):11448–11453. doi: 10.1073/pnas.0404220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Penesyan A, Marshall-Jones Z, Holmstrom C, Kjelleberg S, Egan S. Antimicrobial activity observed among cultured marine epiphytic bacteria reflects their potential as a source of new drugs. FEMS Microbiol Ecol. 2009;69(1):113–124. doi: 10.1111/j.1574-6941.2009.00688.x. [DOI] [PubMed] [Google Scholar]
- 6.Yang Y, Wang J, Liao J, Xie S, Huang Y. Abundance and diversity of soil petroleum hydrocarbon-degrading microbial communities in oil exploring areas. Appl Microbiol Biotechnol. 2015;99(4):1935–1946. doi: 10.1007/s00253-014-6074-z. [DOI] [PubMed] [Google Scholar]
- 7.Holmes NA, Innocent TM, Heine D, Bassam MA, Worsley SF, Trottmann F, Patrick EH, Yu DW, Murrell JC, Schiott M, et al. Genome analysis of two Pseudonocardia Phylotypes associated with Acromyrmex leafcutter ants reveals their biosynthetic potential. Front Microbiol. 2016;7:2073. doi: 10.3389/fmicb.2016.02073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xue Y, Kong C, Shen W, Bai C, Ren Y, Zhou X, Zhang Y, Cai M. Methylotrophic yeast Pichia pastoris as a chassis organism for polyketide synthesis via the full citrinin biosynthetic pathway. J Biotechnol. 2017;242:64–72. doi: 10.1016/j.jbiotec.2016.11.031. [DOI] [PubMed] [Google Scholar]
- 9.Cao J, Jiang M, Li P, Chu Z. Genome-wide identification and evolutionary analyses of the PP2C gene family with their expression profiling in response to multiple stresses in Brachypodium distachyon. BMC Genomics. 2016;17:175. doi: 10.1186/s12864-016-2526-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horn H, Hentschel U, Abdelmohsen UR. Mining Genomes of Three Marine Sponge-Associated Actinobacterial Isolates for Secondary Metabolism. Genome Announc. 2015;3(5). 10.1128/genomeA.01106-15. [DOI] [PMC free article] [PubMed]
- 11.Medema MH, Fischbach MA. Computational approaches to natural product discovery. Nat Chem Biol. 2015;11(9):639. doi: 10.1038/nchembio.1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu B, Hu B, Wang J, Lan Y, Zhu Y, Dai X, Huang L, Huang Y, Du W. Virgibacillus indicus sp. nov. and Virgibacillus profundi sp. nov, two moderately halophilic bacteria isolated from marine sediment by using microfluidic streak plates. Int J Syst Evol Microbiol. 2018;68(6):2015–2023. doi: 10.1099/ijsem.0.002782. [DOI] [PubMed] [Google Scholar]
- 13.Tak EJ, Kim HS, Lee JY, Kang W, Sung H, Kim PS, Hyun DW, Shin NR, Roh JR, Park SD, et al. Virgibacillus phasianinus sp. nov., a halophilic bacterium isolated from faeces of a Swinhoe's pheasant, Lophura swinhoii. Int J Syst Evol Microbiol. 2018;68(4):1190–1196. doi: 10.1099/ijsem.0.002650. [DOI] [PubMed] [Google Scholar]
- 14.Oh YJ, Jang JY, Lim SK, Kwon MS, Lee J, Kim N, Shin MY, Park HK, Seo MJ, Choi HJ. Virgibacillus kimchii sp. nov., a halophilic bacterium isolated from kimchi. J Microbiol. 2017;55(12):933–938. doi: 10.1007/s12275-017-7386-3. [DOI] [PubMed] [Google Scholar]
- 15.Montriwong A, Rodtong S, Yongsawatdigul J. Detergent-stable salt-activated proteinases from Virgibacillus halodenitrificans SK1-3-7 isolated from fish sauce fermentation. Appl Biochem Biotechnol. 2015;176(2):505–517. doi: 10.1007/s12010-015-1591-5. [DOI] [PubMed] [Google Scholar]
- 16.Toopcham T, Mes JJ, Wichers HJ, Yongsawatdigul J. Immunomodulatory activity of protein hydrolysates derived from Virgibacillus halodenitrificans SK1-3-7 proteinase. Food Chem. 2017;224:320–328. doi: 10.1016/j.foodchem.2016.12.041. [DOI] [PubMed] [Google Scholar]
- 17.Kumar S, Karan R, Kapoor S, PS S, KK S. Screening and isolation of halophilic bacteria producing industrially important enzymes. Braz J Microbiol. 2012;43(4):1595–1603. doi: 10.1590/S1517-83822012000400044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Phrommao E, Yongsawatdigul J, Rodtong S, Yamabhai M. A novel subtilase with NaCl-activated and oxidant-stable activity from Virgibacillus sp. SK37. BMC Biotechnol. 2011;11:65. doi: 10.1186/1472-6750-11-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sinsuwan S, Rodtong S, Yongsawatdigul J. Purification and characterization of a salt-activated and organic solvent-stable heterotrimer proteinase from Virgibacillus sp. SK33 isolated from Thai fish sauce. J Agric Food Chem. 2010;58(1):248–256. doi: 10.1021/jf902479k. [DOI] [PubMed] [Google Scholar]
- 20.Al-Amoudi S, Essack M, Simões MF, Bougouffa S, Soloviev I, Archer JA, Lafi FF, Bajic VB. Bioprospecting Red Sea coastal ecosystems for Culturable microorganisms and their antimicrobial potential. Marine Drugs. 2016;14(9):165. doi: 10.3390/md14090165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Al-Amoudi S, Razali R, Essack M, Amini MS, Bougouffa S, Archer JA, Lafi FF, Bajic VB. Metagenomics as a preliminary screen for antimicrobial bioprospecting. Gene. 2016;594(2):248–258. doi: 10.1016/j.gene.2016.09.021. [DOI] [PubMed] [Google Scholar]
- 22.Yeong M. BiG-SCAPE: exploring biosynthetic diversity through gene cluster similarity networks. 2016. [Google Scholar]
- 23.Luo C, Liu X, Zhou H, Wang X, Chen Z. Nonribosomal peptide synthase gene clusters for lipopeptide biosynthesis in Bacillus subtilis 916 and their phenotypic functions. Appl Environ Microbiol. 2015;81(1):422–431. doi: 10.1128/AEM.02921-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klemetsen T, Raknes IA, Fu J, Agafonov A, Balasundaram SV, Tartari G, Robertsen E, Willassen NP. The MAR databases: development and implementation of databases specific for marine metagenomics. Nucleic Acids Res. 2018;46(D1):D692–D699. doi: 10.1093/nar/gkx1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Othoum G, Bougouffa S, Razali R, Bokhari A, Alamoudi S, Antunes A, Gao X, Hoehndorf R, Arold ST, Gojobori T, et al. In silico exploration of Red Sea Bacillus genomes for natural product biosynthetic gene clusters. BMC Genomics. 2018;19(1):382. doi: 10.1186/s12864-018-4796-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hyatt D, Chen G-L, LoCascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;11(1):119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alam I, Antunes A, Kamau AA, Kalkatawi M, Stingl U, Bajic VB. INDIGO–INtegrated data warehouse of MIcrobial GenOmes with examples from the red sea extremophiles. PLoS One. 2013;8(12):e82210. doi: 10.1371/journal.pone.0082210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lex A, Gehlenborg N, Strobelt H, Vuillemot R, Pfister H. UpSet: visualization of intersecting sets. IEEE Trans Vis Comput Graph. 2014;20(12):1983–1992. doi: 10.1109/TVCG.2014.2346248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khan A, Mathelier A. Intervene: a tool for intersection and visualization of multiple gene or genomic region sets. BMC Bioinformatics. 2017;18(1):287. doi: 10.1186/s12859-017-1708-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bertelli C, Laird MR, Williams KP, Simon Fraser University Research Computing G. Lau BY, Hoad G, Winsor GL, FSL B. IslandViewer 4: expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res. 2017;45(W1):W30–W35. doi: 10.1093/nar/gkx343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Langille MG, Brinkman FS. IslandViewer: an integrated interface for computational identification and visualization of genomic islands. Bioinformatics. 2009;25(5):664–665. doi: 10.1093/bioinformatics/btp030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carver T, Thomson N, Bleasby A, Berriman M, Parkhill J. DNAPlotter: circular and linear interactive genome visualization. Bioinformatics. 2008;25(1):119–120. doi: 10.1093/bioinformatics/btn578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Emms DM, Kelly S. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 2015;16(1):157. doi: 10.1186/s13059-015-0721-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44(W1):W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weber T, Blin K, Duddela S, Krug D, Kim HU, Bruccoleri R, Lee SY, Fischbach MA, Müller R, Wohlleben W. antiSMASH 3.0—a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 2015;43(W1):W237–W243. doi: 10.1093/nar/gkv437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cimermancic P, Medema MH, Claesen J, Kurita K, Brown LCW, Mavrommatis K, Pati A, Godfrey PA, Koehrsen M, Clardy J. Insights into secondary metabolism from a global analysis of prokaryotic biosynthetic gene clusters. Cell. 2014;158(2):412–421. doi: 10.1016/j.cell.2014.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Network visualization of 35 gene cluster families in Virgibacillus, showing that few groups are found in the majority of Virgibacillus genomes and none are found in all nine genomes (DOCX 100 kb)
Table S1. Features of modular clusters identified in Virgibacillus genomes (DOCX 16 kb)
Table S2. Homology analysis of the modular genes in six clusters identified in Red Sea Virgibacillus strains to marine metagenomes. (DOCX 13 kb)
Data Availability Statement
All data used in this study have been included in this article.