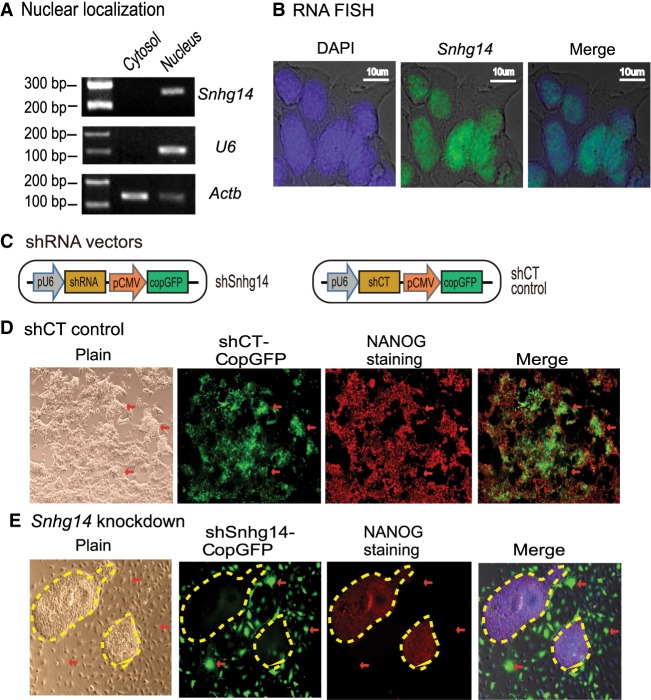
Figure 5.
Sox2-interacting lncRNAs are important in maintaining pluripotency. (A) Subcellular localization of Snhg14 lncRNA. Cytoplasmic and nuclear RNAs were isolated, and RT-PCR was used to quantitate the subcellular distribution of Snhg14. Actb was used as the cytoplasmic control and U6 was used as the nuclear control. (B) RNA-FISH of Snhg14. To detect the mature RNA, the DIG-11-dUTP probe was synthesized as a short single-strand DNA to cover the intron splicing site. The Snhg14 signal was detected by anti-digoxigenin-fluorescein (green). DAPI was used to stain the nucleus of iPSC (blue). Snhg14 was predominantly located in the nucleus. (C) The shRNA knockdown vectors. The CopGFP in the lentiviral vector was used as the marker to track the shRNA knockdown cells. (D) The iPSCs transfected with shRNA control (shCT). Pluripotency was examined by immunohistochemical staining of critical stem cell marker NANOG protein. The lentivirus-transfected iPSCs expressed the CopGFP fluorescence marker (red arrow) and still stained positive for the pluripotency marker NANOG (red). These cells maintained the original cell morphology. (E) The Snhg14 knockdown iPSCs. (Red arrow) The shRNA-knocked down cells that show the exit from pluripotency, with enlarged and flat cell morphology. The Snhg14 knockdown iPSCs lost the NANOG pluripotency marker. The yellow-marked areas represent the “island” cells that escaped lentiviral infection. They maintained the compact stem cell appearance and stained positive for the NANOG marker.