Figure 1.
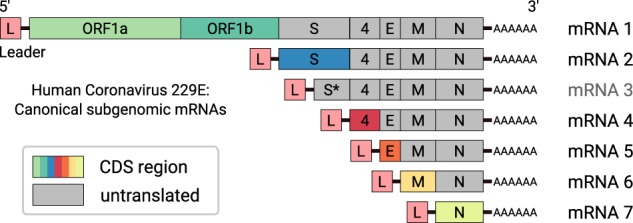
Scheme of genomic and subgenomic (sg) RNAs produced in HCoV-229E–infected cells (Raabe et al. 1990; Schreiber et al. 1989). Translation of the 5′-terminal ORF(s) of the respective mRNA gives rise to the various viral structural and nonstructural proteins (indicated by different colors). mRNA 3 is considered defective and unlikely to be translated into protein. Each mRNA has a 3′ poly(A) tail and carries a 5′-leader sequence that is identical to the 5′-end of the genome. In a process called discontinuous extension of negative strands, negative-strand RNA synthesis is attenuated at specific genomic positions. After transfer of the nascent strand to an upstream position on the template, RNA synthesis is continued by copying the 5′-leader sequence. As a result, the 3′-ends of coronavirus minus-strand mRNAs are equipped with the complement of the 5′-leader. The latter process is guided by base-pairing interactions between the transcription-regulating sequence (TRS) located immediately downstream from the leader sequence (TRS-L) at the 5′-end of the genome and the sequence complement of a TRS located upstream of one of the ORFs present in the 3′-proximal genome region (TRS-B).
