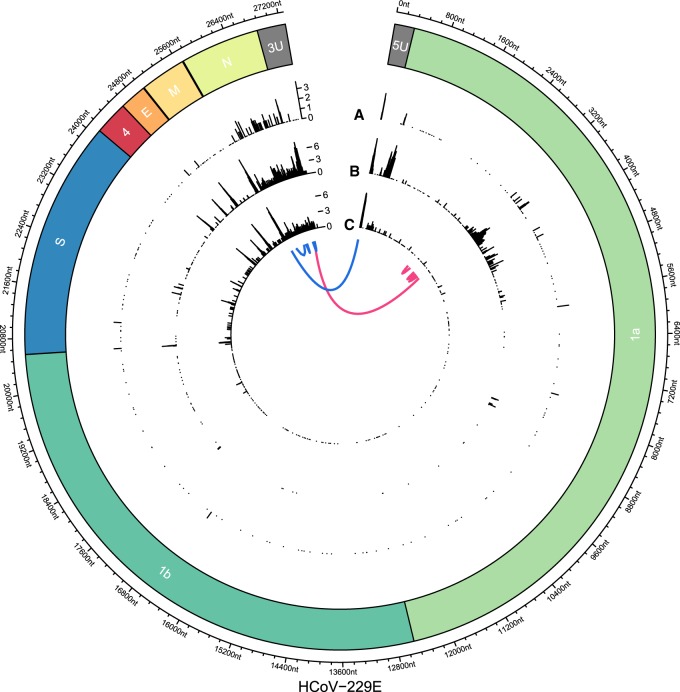
Figure 3.
Joining of noncontiguous genome sequences in sg RNAs identified in HCoV-229E–infected cells. On the circular axis, the annotations of the reference genome (including 5′ UTR [5U] and 3′ UTR [3U]) are shown. Genomic positions of “discontinuous sites” identified in Illumina reads (A; outer track), nanopore reads of sample HCoV-229E WT (B; middle track), and nanopore reads of SL2 sample (C; inner track) reveal multiple recombination sites across the whole genome. An aggregation of recombination sites can be observed in the region that encodes the viral N protein. Furthermore, clear recombination sites can be seen at intergenic boundaries and at the 5′ and 3′ UTRs, with the former corresponding to the boundary between the leader sequence and the rest of the genome. Another prominent cluster can be observed in ORF1a in the WT nanopore sample but not in SL2. This cluster is supported by the WT Illumina data, excluding sequencing bias as a potential source of error. We hypothesize that because samples WT and SL2 were obtained from nonplaque-purified serially passaged virus populations derived from in vitro transcribed genome RNAs transfected into Huh7 cells, differences in the proportion of full-length transcripts versus abortive transcripts could translate into different patterns of recombination. Generally, nanopore-based sequencing allows more detailed analysis of recombination events owing to the long read length. Even complex isoforms such as two exemplary reads, each with four discontinuous segments, can be observed (blue and pink).