Figure 1.
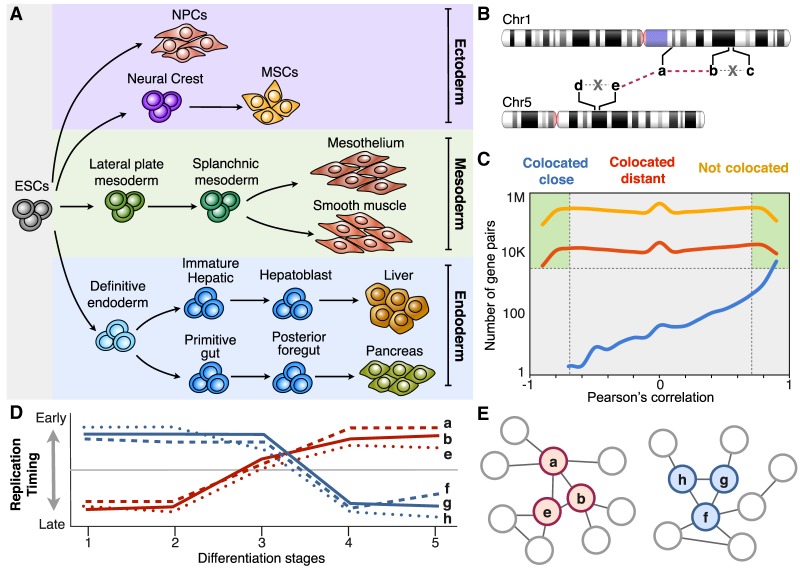
Coordinated changes in RT can be exploited to construct gene regulatory networks. (A) RT programs of distinct cell types representing intermediate stages of human embryonic stem cell differentiation toward endoderm, mesoderm, and ectoderm were analyzed for the construction of RT regulatory networks. (B) Depiction of different highly RT-correlated genes from distinct chromosomes and the establishment of network interaction edges between them. From all possible combinations of gene pairs, those colocated within 500 kb were removed from the analysis to include exclusively distal gene interactions. Regulatory interactions (edges) between gene pairs are considered only for genes located >500 kb apart (colocated distant) or in distinct chromosomes (not colocated); that is, edges between genes b-c and d-e were not included in the analysis. (C) Number of gene pairs as function of RT correlation for distinct categories of gene pairs: colocated close (within 500 kb), colocated distant (separated by >500 kb), and not colocated (from different chromosomes). Only gene pairs with significant RT correlations and located at least 500 kb apart were considered. (D) RT patterns of distinct hypothetical genes during five distinct differentiation intermediate stages (distinct number of differentiation intermediates were used from each pathway). (E) Construction of RT regulatory networks based on the significant Pearson's correlation (distance between nodes are proportional to the correlation strength).