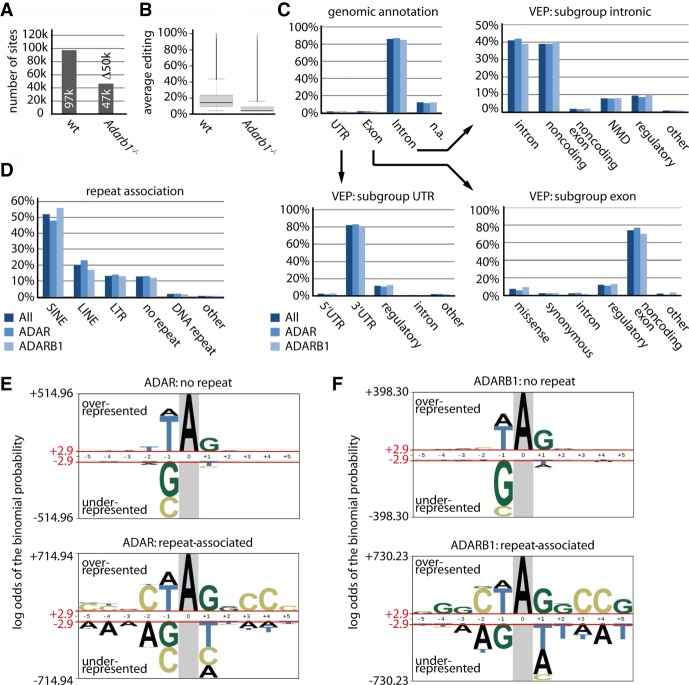
Figure 2.
Both ADAR- and ADARB1-mediated editing is primarily associated with intronic regions and repeat elements. (A,B) Adarb1 deletion leads to an ∼60% reduction in editing activity. (A) The total number of editing sites is shown separately for wild type and Adarb1−/− (sites edited by ADAR). (B) The average editing level of all identified editing sites is plotted for wild type and Adarb1−/− (overall ADAR editing activity). Error bars = standard deviation. n = 3. (C) The genomic annotation for all sites (dark blue), ADAR sites (blue), and ADARB1 sites (light blue) is given (Exon, Intron, UTR, [n.a.] not annotated/intergenic). In addition, for the subgroups Exon, Intron, and UTR, the predicted effect of editing is given using Ensembl's variant effect predictor (VEP). VEP terms: intron variant (intron); noncoding transcript variant (noncoding); noncoding transcript exon variant (noncoding exon); NMD transcript variant (NMD); regulatory region (regulatory); 5′ or 3′ UTR variant (5′ or 3′ UTR); missense variant (missense); synonymous variant (synonymous). (D) The percentage of editing sites associated with a particular repeat as identified by RepeatMasker is shown. Colors as in C. (E,F) Sequences enriched close to editing sites not associated with repeat elements (upper panel) or associated with repeat elements (lower panel) depicted separately for (E) ADAR or (F) ADARB1. The height of the nucleotide indicates either the degree of overrepresentation (above the line) or the degree of underrepresentation (below the line).