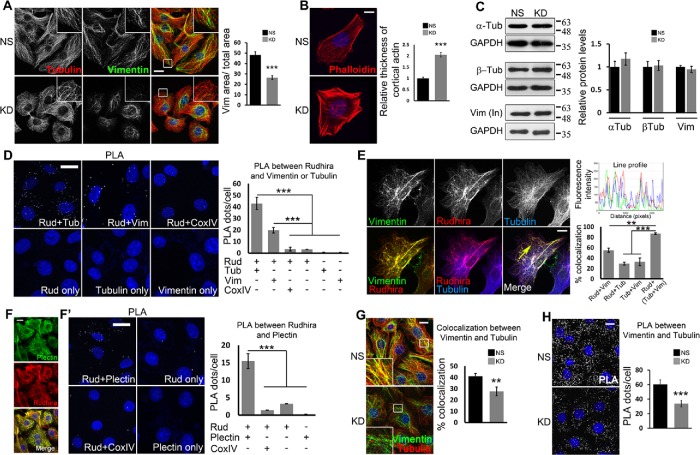
FIGURE 1:
Rudhira interacts with and controls cross-talk between MTs and IFs. (A, B) NS and KD cells were costained for cytoskeleton markers, tubulin and vimentin (A) or phalloidin (B) to detect gross cytoskeleton organization. Boxed regions are magnified in the insets. The graph in A shows the ratio of vimentin IF area to total cell area (11 cells), and the graph in B shows the relative thickness of cortical actin (25 cells). (C) Immunoblots detect the levels of cytoskeletal proteins. Graphs show the quantitation of relative protein levels. (D) Direct interaction between Rudhira and tubulin or vimentin in wild-type SVECs analyzed by PLA. Cells stained with a single antibody or the Rudhira+CoxIV combination were taken as negative controls. The graph shows the quantitation of PLA dots per cell indicating the extent of interaction. (E) Relative localization of vimentin IFs, Rudhira, and MTs was performed by triple immunostaining in wild-type SVECs. The line profile shows the fluorescence intensity peaks for the three colors along the yellow arrow. The graph shows the percent overlap of Rudhira with vimentin or tubulin or vimentin+tubulin pattern, as indicated (15 cells). (F, F’) Relative association of Rudhira with the cytolinker plectin by immunofluorescence (F) or PLA (F’). Cells stained with a single antibody or Rudhira+CoxIV combination were taken as negative controls. The graph shows the quantitation of PLA dots per cell indicating the extent of interaction. (G) NS or KD cells were analyzed for coalignment of vimentin and MTs by coimmunofluorescence. Boxed regions are magnified in the insets. The graph shows the percent overlap between vimentin and tubulin patterns (30 cells). (H) Vimentin and MT association by PLA. The graph shows the quantitation of PLA dots per cell indicating extent of interaction. Error bars indicate SEM. Results shown are a representative of at least three independent experiments with at least three biological replicates. Statistical analysis was carried out using one-way ANOVA. Scale bar: (A, D, F, F’, G, H) 10 μm, (B, E) 20 μm. **p < 0.01, ***p < 0.001.