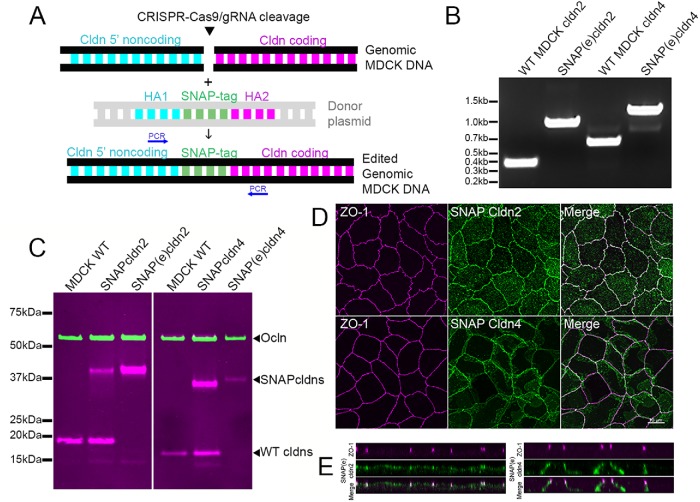
FIGURE 3:
CRISPR/Cas9 gene editing was used to insert SNAP tags at the N-terminal ends of cldns2 and 4. (A) Scheme for insertion of SNAP tags into MDCK cell DNA; CRISPR/Cas9/gRNA was used to cleave DNA specifically upstream of coding regions and homology-directed recombination was used to insert SNAP tag; cells were labeled with fluorescent SNAP tag ligand and single cells sorted into multiwell dishes. (B) Positive clones were identified by PCR (position of primers indicated in A) and by immunoblot (C), where expression was compared with wild-type MDCK II cells and cells stably overexpressing SNAP-cldns (left panel probed for ocln and cldn2, right for ocln and cldn4). The endogenously tagged cldn4 fusion protein contains five extra residues between the SNAP tag and the start of cldn4, resulting in a slightly larger fusion protein. Ocln was used as a loading control. The upper magenta band in the cldn4 panel (overlapping with and slightly above ocln) is a nonspecific band seen with lots of IR-labeled secondary antibodies. (D) Maximum intensity projections of confocal images of ZO-1 colocalized JF549 SNAP-ligand labeled SNAP(e)cldn2 (top panels) and SNAP(e)cldn4 (bottom panels) shows localization like that of endogenous cldns, as do z-view images (E).