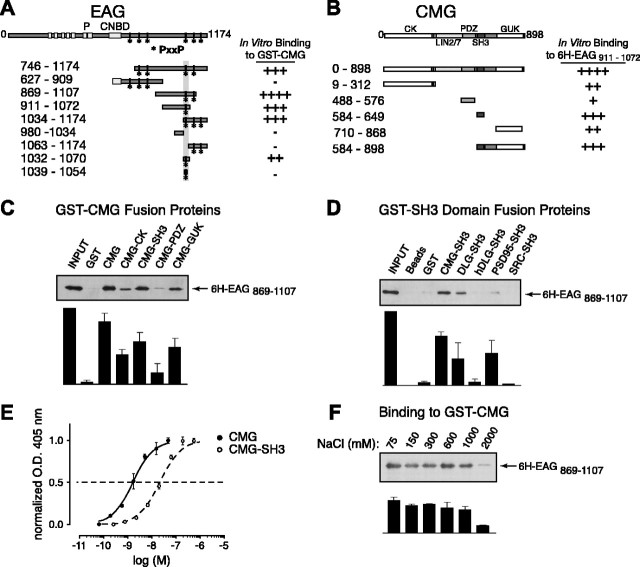
Figure 5.
A direct interaction between EAG and CMG adaptor protein in in vitro binding assays. A, Schematic representation and respective binding for the EAG protein constructs used in mapping experiments. For comparison, full-length EAG is shown at the top with the primary domains as indicated, including the six putative transmembrane domains, pore (P), and the region with homology to cyclic nucleotide-binding domains (CNBD). The amino acid residues contained in each EAG construct are noted by the subscripted numbers on the left, and the relative binding of each construct to full-length CMG is indicated on the right. Note that the EAG C-terminal cytoplasmic domain contains six SH3-binding motifs as defined by the PxxP consensus sequence. The locations of these potential binding sites are indicated by asterisks. B, Schematic representation and respective binding for the CMG constructs used in mapping experiments as described for A. CK, CaMKII-like domain. Binding to 6H-EAG869-1107 is shown in C-F. This fragment was chosen for display because there was minimal degradation during purification. C, 6H-EAG869-1107 binding to full-length CMG and CMG domains. GST-tagged CMG fragments immobilized on glutathione Sepharose beads were assayed for their ability to pull down 6H-EAG869-1107 as described in Materials and Methods. Top, Western blot probed with MRGS-6H antibody to detect interacting EAG fragments. Bottom, Bound 6H-EAG869-1107 quantified by densitometry; data are presented as the mean ± SEM; n = 3. D, 6H-EAG869-1107 binding to GST-SH3 domain fusion proteins. Top, The ability of purified GST-SH3 domains, derived from the proteins indicated above each lane, to pull down 6H-EAG869-1107 was determined in immunoblots by comparing the relative amount of associating 6H-EAG869-1107 to the amount available during the interaction experiment (left lane). As shown in lanes 2 and 3, there was little or no background binding to either glutathione Sepharose or immobilized GST tag. Bottom, Bound 6H-EAG869-1107 quantified by densitometry; data are presented as the mean ± SEM; n = 3. E, Dose-response curves comparing the binding of CMG and the CMG SH3 domains to 6H-EAG869-1107. Wells were coated overnight with 6H-EAG869-1107, washed, and then incubated with the indicated concentrations of GST fusion protein. Bound protein was detected using anti-GST antibody, followed by alkaline phosphatase-conjugated secondary antibody and treatment with p-nitrophenylphosphate. Binding was determined by colorimetric reaction at 405 nm. Each concentration was assayed in quadruplicate. Data are presented as the mean ± SEM; n = 4. O.D., Optical density. F, Effect of salt concentration on the 6H-EAG869-1107-CMG complex. Experiments examining the binding to GST-CMG were performed as described in Materials and Methods, with the exception that PBS buffer was modified to include the indicated NaCl concentrations. Complex formation was relatively unaffected in up to 1000 mm salt. Bottom, Bound 6H-EAG869-1107 quantified by densitometry; data are presented as the mean ± SEM; n = 3.