Figure 7.
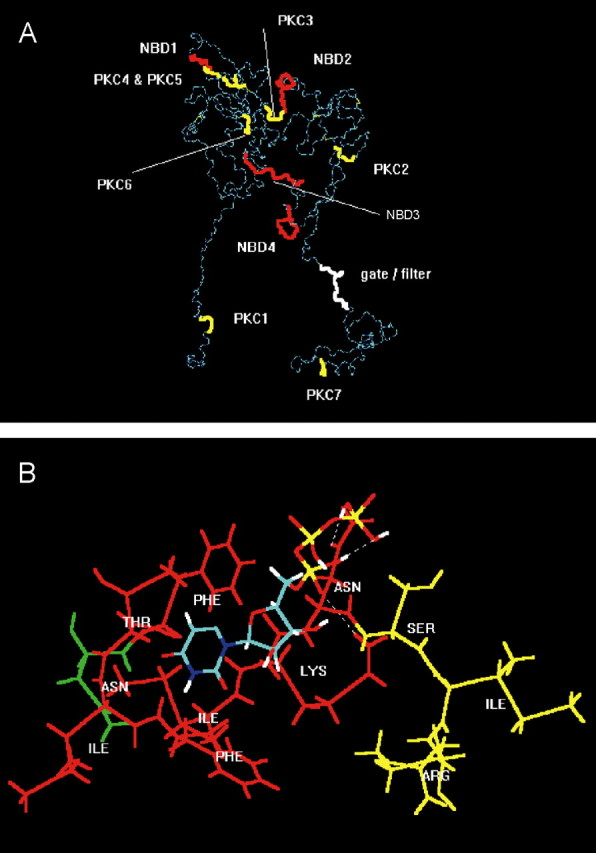
Molecular simulation of nucleotide binding sites and consensus PKC phosphorylation sites at the P2X3 receptor. A, For visual clarity, the backbone structure (in dark and light blue) of the geometry optimized, monomeric, charged, membrane-embedded, unoccupied human P2X3 glycoprotein receptor is shown. The structure is viewed parallel to the plane of a membrane. A five-disulfide core (thin yellow lines) has been assumed. The extracellular domain forms a twisted loop that is solvent dependent. The intracellularly occurring N- and C-terminal PKC phosphorylation sites (thick yellow) are denoted by PKC1 (amino acids in positions 12-14) and PKC7 (365-367); the extracellular PKC phosphorylation sites are denoted by PKC2-PKC6 (134-136, 178-180, 196-198, 202-204, and 269-271). The extracellularly occurring potential NBDs (red) are denoted by NBD1-NBD4 (62-66, 171-177, 279-286, and 295-302). The potential filter (white) is probably formed by the residues in positions 333-341. The fully functional receptor is a 3n homomeric (n = 1, 2,...) ion channel. B, Model of a putative interaction of UTP (colors: green, carbon; red, oxygen; white, hydrogen; blue, nitrogen; yellow, phosphorus) with the second putative nucleotide binding domain NBD2 of human P2X3 (Phe-Thr-Ile-Phe-Lys-Asn, 171-177; red), which is in steric neighborhood of the PKC phosphorylation site PKC3 (amino acids Ser-Ile-Arg in positions 178-180; yellow). The first residue (Asn, green) of the N-glycosylation attachment site (Asn-Phe-Thr-Ile, 170-173) is also shown. The uridine ring of UTP is bound into a lipophilic pocket (Phe171, Ile175); the oxygen atoms of the phosphate groups form hydrogen bonds with the hydrogen donating groups of the residues of Lys-176 and Asn-177.
