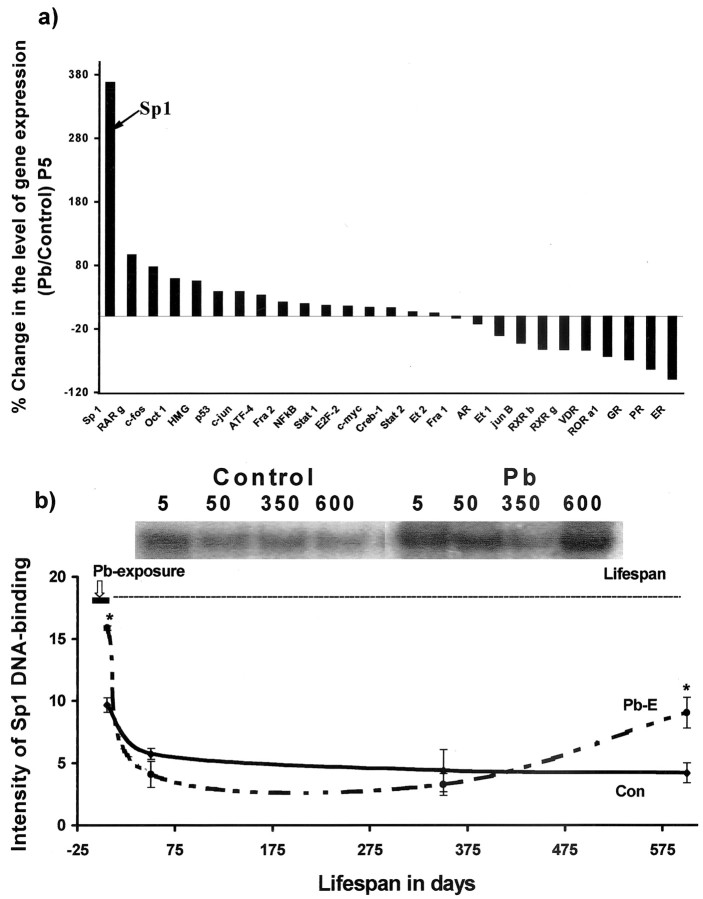
Figure 2.
mRNA profiling of transcription factors after developmental exposure to Pb and lifetime changes in Sp1 DNA-binding in the cortices of control and Pb-exposed animals. a, mRNA profiling. Cortical total RNA was isolated from P5 rats and used to probe cDNA arrays of transcription factors: AR, androgen receptor; ATF-4, activating transcription factor-4; c-fos, c-jun, c-myc, oncogenes; Creb-1, cAMP-responsive element-binding protein-1; E2F-2, ubiquitin-conjugating enzyme F-2; ER, estrogen receptor; Et, erythroblastosis virus oncogene homology; GR, glucocorticoid receptor; HMG, high-mobility group protein; NFκB, nuclearfactor κB; Oct 1, octamer transcription factor 1; p53, phosphoprotein 53; PR, progesterone receptor; RAR, retinoic acid receptor [related factors: retinoid orphan receptor (ROR), retinoid X receptor (RXR)]; Stat, signal transducer and activator of transcription; VDR, vitamin D3 receptor. The array data shown were derived from two sets of experiments, and each set consisted of tissue pooled from three animals. b, Sp1 DNA-binding. Nuclear extracts were prepared from controls and animals exposed as indicated by the bar in the figure. Sp1 DNA-binding was analyzed using gel mobility shift assays. Sp1 DNA-binding data shown are derived from two cohorts of animals separated in time by 1 year; each data point represents the mean ± SEM for four to six animals. Data were analyzed by two-way ANOVA followed by a Duncan's post hoc test to compare the effects among various treatments. Values marked with an asterisk are significantly different from their corresponding controls (p < 0.05).