Figure 2.
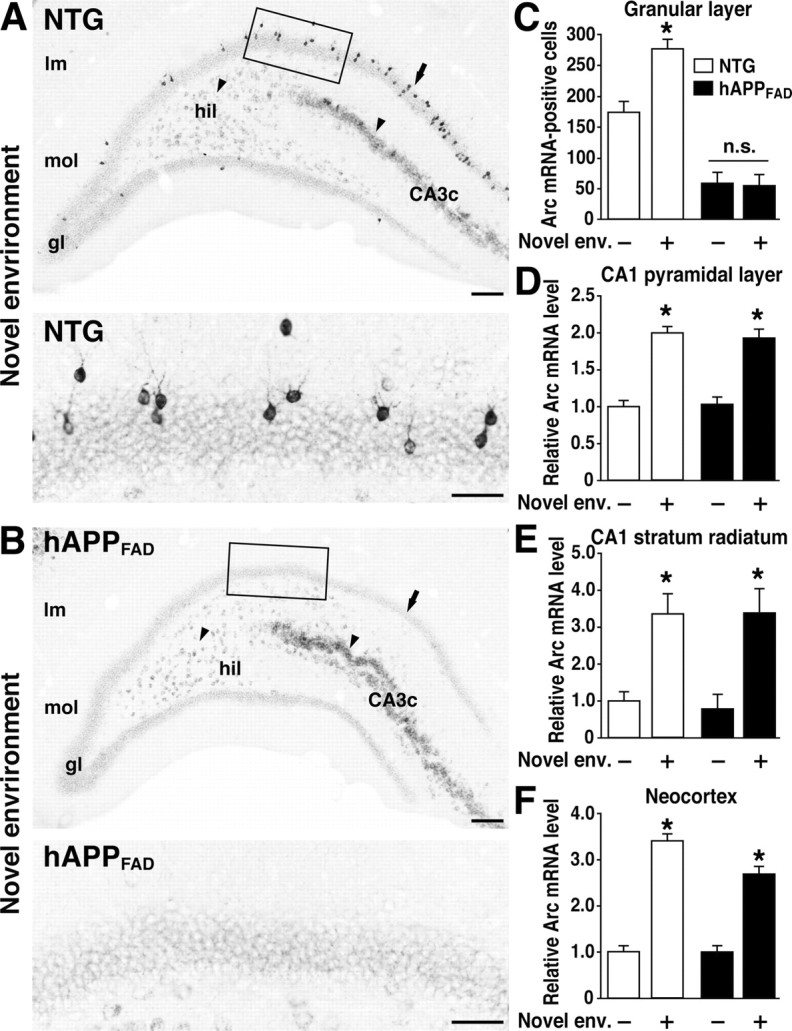
Lack of Arc induction in granule cells of hAPPFAD mice after exploration of a novel environment. A, B, In situ hybridization illustrates the heterogeneity of Arc-expressing cells in the dentate gyrus and the differential susceptibility of distinct neuronal populations to impairments in Arc expression in hAPPFAD mice. Note the induced Arc expression in granule cells (arrow), hilar cells (arrowhead), and proximal CA3c pyramidal cells (arrowhead) in an NTG mouse (A) and the selective absence of Arc mRNA induction in the granule cells (arrow) of an hAPPFAD mouse (B). The bottom panels depict higher-magnification images from the squared regions in A and B. Scale bars: top, 100 μm; bottom, 50 μm. lm, Stratum lacunosum-moleculare; mol, molecular layer; gl, granular layer; hil, hilus. C-F, Quantitation of Arc expression at baseline and after exploration of a novel environment (Novel env.). The number of Arc-positive granule cells and the optical density of in situ hybridization labels in other brain regions were assessed in coronal sections from NTG controls (□) and hAPPFAD mice (▪) that had (+) or had not (-) explored a novel environment (n = 11-12 per genotype and condition). hAPPFAD mice had significantly fewer Arc-positive cells in the granular layer of the dentate gyrus than NTG controls, both at baseline and after exploration (C). Basal levels of Arc mRNA in the CA1 pyramidal layer (D), CA1 stratum radiatum (E), and neocortex (F) were similar in hAPPFAD mice and NTG controls. In both groups, Arc mRNA expression in these regions increased robustly after exploration of the novel environment. *p < 0.01 versus home-caged group of the same genotype (Tukey-Kramer test). Error bars indicate SEM. n.s., Nonsignificant.
