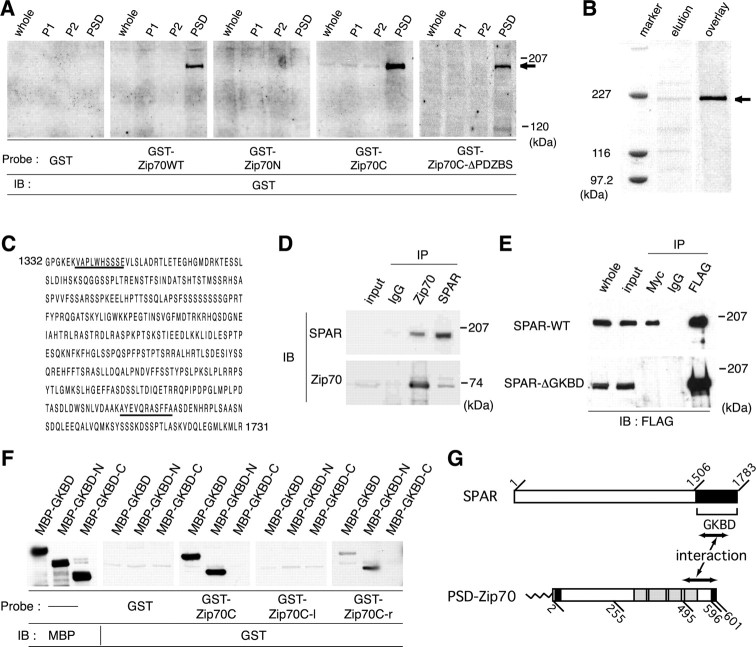
Figure 7.
Identification of SPAR as the PSD-Zip70 binding partner. A, Ligand overlay binding assay to identify PSD-Zip70 binding partner(s). The indicated GST-fusion proteins were used as ligands. Bound GST-fusion proteins were visualized by immunoblotting (IB). The arrow indicates a 200 kDa PSD protein. Whole, P1, S2, and PSD indicate the whole-brain lysate, crude membrane, soluble, and PSD fractions, respectively. B, Partial purification of a 200 kDa PSD protein using a GST-fused PSD-Zip 70C affinity column. A partially purified 200 kDa PSD protein (arrow) was stained with Coomassie brilliant blue (elution) and detected by ligand overlay assay using GST-fused PSD-Zip 70C as a ligand (overlay). C, Aminoacid sequences of the SPAR C-terminal region. Underlining indicates partial sequences of two peptides from the 200 kDa PSD protein. D, Coimmunoprecipitation of PSD-Zip70 and SPAR from the brain extracts. The brain extracts were coimmunoprecipitated (IP) with anti-PSD-Zip70 or anti-SPAR antibodies or normal rabbit IgG and were analyzed by immunoblotting (IB) for SPAR or PSD-Zip70. E, In vitro interaction between PSD-Zip70 and SPAR. The COS-7 cell lysates cotransfected with PSD-Zip70-Myc and FLAG-SPAR-WT or FLAG-SPAR-ΔGKBD were immunoprecipitated (IP) with anti-Myc or anti-FLAG antibodies and analyzed by immunoblotting (IB) for FLAG. F, Ligand overlay binding assay to determine the binding domains of PSD-Zip70 and SPAR. GKBD, GKBD-N, and GKBD-C indicate the regions corresponding to residues 1583-1783, 1584-1714, and 1715-1783 of SPAR, respectively. Zip70C, Zip70C-l, and Zip70C-r indicate the regions corresponding to residues 246-601, 246-494, and 495-601 of PSD-Zip70, respectively. GST, GST-Zip70C, GST-Zip70C-l, and GST-Zip70C-r were used as ligands. GKBD-C and GKBD-N indicate the C-terminal and N-terminal halves of SPAR-GKBD. G, Schematic diagram of the interaction between PSD-Zip70 and SPAR.