FIG A2.
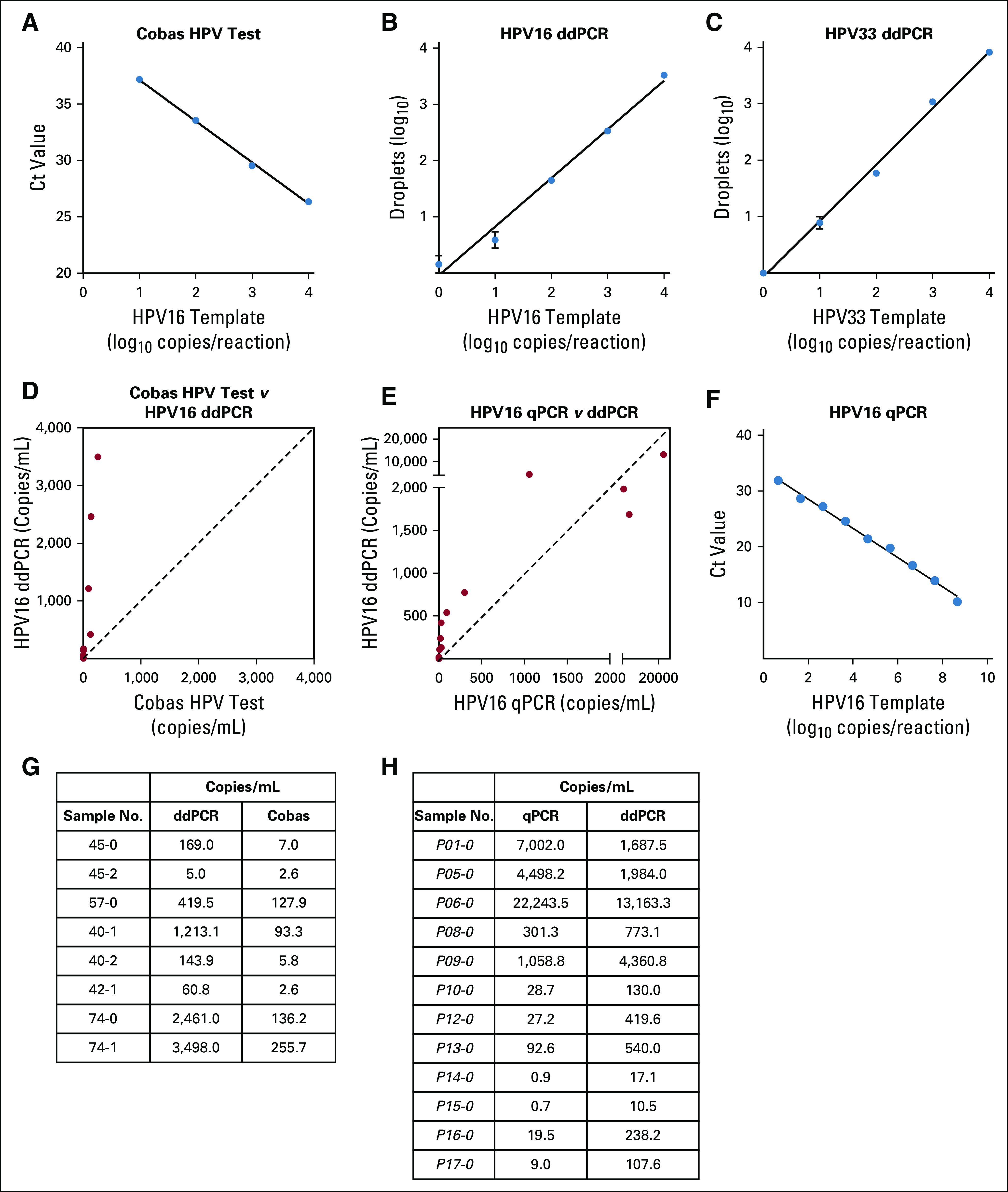
Human papillomavirus (HPV) droplet digital polymerase chain reaction (ddPCR) assay performance relative to established polymerase chain reaction (PCR)–based HPV detection techniques. (A-C and F) Standard curves of Roche Cobas HPV test, HPV16 ddPCR, HPV33 ddPCR, and HPV16 quantitative PCR (qPCR) using a serial dilution of HPV16 plasmid template on a plasmid. (D and G) Comparison of HPV16 ddPCR and the Cobas HPV test in the same eight samples. ddPCR detected a higher number of copies per milliliter of plasma (P = .008, Wilcoxon matched-pairs signed rank test), whereas circulating tumor DNA (ctDNA) levels detected by both methods are highly correlated (r2 = 0.82, P = .002). The signal in paired samples was a median of 15.9-fold higher with ddPCR. The proprietary primer and probe sequences of the Roche Cobas test result in an amplicon length of approximately 200 base pairs (bp), likely too large for fragmented ctDNA, which has a median size of 176 bp.9 (E and H) Comparison of HPV16 ddPCR and HPV16 qPCR in the same 12 samples. We found ddPCR to be more sensitive at lower ctDNA values (arbitrary cutoff, less than 1,000 HPV ctDNA copies/mL) by Wilcoxon matched-pairs signed rank test (P = .008), although each approach was highly correlated (r2 = 0.88, P < .001). Among all paired samples, HPV ctDNA was a median of 5.2-fold higher with ddPCR. At high levels of HPV16, the HPV ctDNA signal can begin to saturate, as observed in three samples because only 20,000 droplets are read on the Bio-Rad QX200 platform. The higher sensitivity of the ddPCR assay despite a larger amplicon size may be secondary to the partitioning of DNA into droplets and reduced inhibitory factors compared with qPCR29 Ct, cycle threshold.
