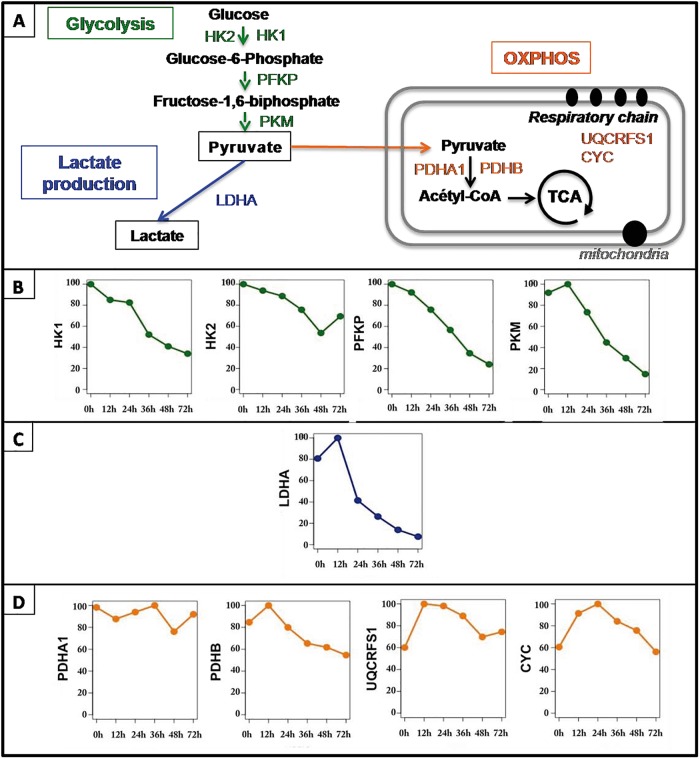
Fig 1. Relevant proteins for glycolytic metabolism and OXPHOS analysis.
A: Proteins were selected from three different categories: key enzymes involved in glycolysis regulation (Glycolysis), LDHA alone, and OXPHOS-related enzymes (OXPHOS). B-D: Protein quantification was performed using mass spectrometry (MS/MS) upon three independent self-renewing T2EC populations and three independent T2EC differentiation kinetics (T2EC induced to differentiate for 12, 24, 36, 48 and 72h). Mean protein levels were normalized as follows: for each enzyme the maximal value was considered equal to 100, and all other values were proportionally converted. Normalized protein levels are represented on the y-axis whereas x-axis correspond to differentiation kinetic time-points (hours). LDHA: Lactate dehydrogenase A, HK1: Hexokinase isoform 1, HK2: Hexokinase isoform 2, PFKP: Phosphofructokinase Platelet, PKM: Pyruvate Kinase Muscle, PDHA1: Pyruvate Dehydrogenase E1 Alpha 1 Subunit, PDHB: Pyruvate Dehydrogenase E1 Beta Subunit, UQCRFS1: Ubiquinol-Cytochrome C Reductase, Rieske Iron-Sulfur Polypeptide 1, CYC: Cytochrome C.