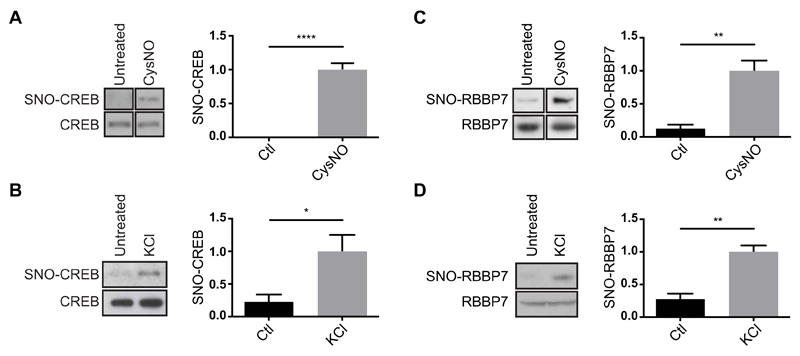
Fig. 2. S-nitrosylation of CREB and RBBP7 in cortical neurons.
(A) E17 rat cortical neurons were cultured for 4 days and treated with CysNO (200 μM for 20 min). SNO-Ps were isolated by SNORAC (n = 2 experiments) or biotin switch (n = 2). Noncontiguous lanes from the same experiment and blot are shown side by side. Ctl, control. (B) Cortical neurons were exposed to KCl (50 mM for 20 min), and SNO-Ps were isolated by SNORAC (n = 2) or biotin switch (n = 1). Data were combined for analysis. (C) Neurons were treated with CysNO (200 μM for 20 min), and SNO-Ps were isolated by biotin switch (n = 4 experiments). Non-contiguous lanes from the same experiment and blot are shown side by side. (D) Cortical neurons were cultured for 4 days and exposed to 50 mM KCl. SNO-Ps were isolated by biotin switch (n = 2 using KCl for 30 min, n = 1 using KCL for 20 min). Data were combined for analysis. For both CREB and RBBP7, isolated proteins were separated by SDS-PAGE, and Western blot analysis was carried out using CREB or RBBP7 antibodies. Densitometry analysis on the Western blots was performed using ImageJ. SNO-P signals were normalized to inputs. Data are means ± SEM. *P ≤ 0.05, **P ≤ 0.01, and ****P ≤ 0.0001 by unpaired t test.