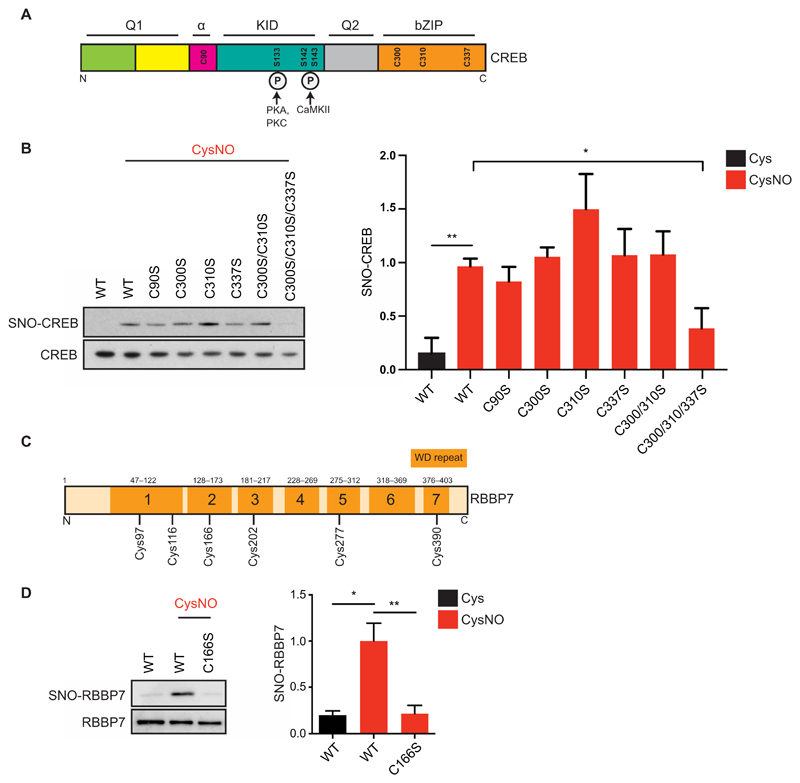
Fig. 4. Identification of CREB and RBBP7 SNO sites.
(A) Domain structure of CREB. Q1, glutamine-rich region 1; α, alpha domain; KID, kinase inducible domain; Q2, glutamine-rich region 2; bZIP, bZIP domain. CREB1 cysteines are annotated (Cys90, Cys300, Cys310, and Cys337). Phosphorylated serines (Ser133, Ser142, and Ser143) are shown along with their respective kinases [protein kinase A (PKA), PKC, and Ca2+/calmodulin-dependent protein kinase II]. (B) Vectors expressing myc-tagged WT CREB (WT) or CREB Cys-to-Ser mutants were transfected into HEK293T cell; 48 hours later, cell lysates were harvested and exposed to Cys or CysNO (500 μM for 20 min) and subjected to the biotin-switch assay (n = 3 experiments). (C) RBBP7 contains seven WD (tryptophan–aspartic acid)–repeat domains, composed of short motifs of about 40 amino acids and ending in a WD dipeptide. (D) Vectors expressing myc-tagged WT RBBP7 (WT) or RBBP7 Cys166-to-Ser mutant (C166S) were transfected into HEK293T cells. After 48 hours, cells were treated with Cys or CysNO (500 μM for 20 min), and lysates were subjected to the biotin-switch assay (n = 3). Isolated proteins and total inputs were separated by SDS-PAGE and then immunoblotted using an antibody against myc. Densitometry analysis was carried out using ImageJ. SNO signals were normalized to total inputs and expressed as fold change relative to WT + CysNO. All data are means ± SEM. *P ≤ 0.05 and **P < 0.01 by one-way ANOVA, compared to WT + CysNO column, and Fisher’s LSD.