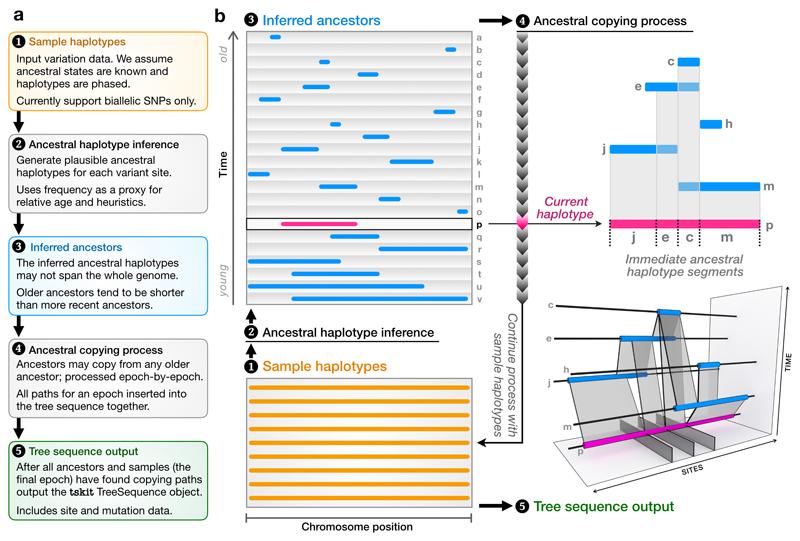
Figure 2.
A schematic of the major steps of the inference algorithm. Starting from a set of sample haplotypes extending over the genome (1), we use the ancestral haplotype inference method (2) to reconstruct fragments of ancestral sequence (3), then infer copying paths among these ancestors (4). The ancestral copying process is shown on the right, using an arbitrary haplotype (p) for illustration. As we move from left to right along p we infer that it has most recently copied from j, e, c and then m. Incorporating the copying history of all older haplotypes (for example, m copied partly from c and partly from h), partial coalescent trees emerge in the bottom-right panel. Once copying paths have been found for all ancestors and samples, we output a tskit tree sequence (5).