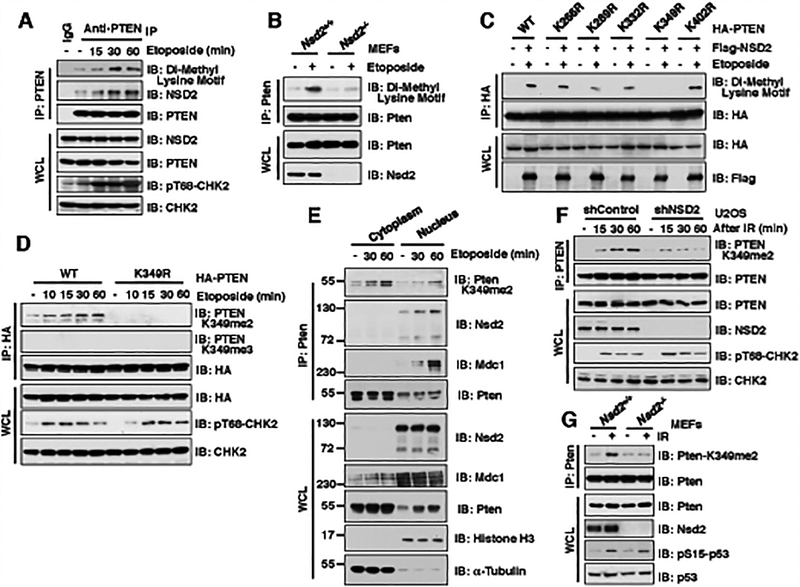
Figure 2. DNA damage promotes NSD2-mediated di-methylation of PTEN at K349.
(A) Methylation of PTEN was detected using the Di-Methyl Lysine motif antibody. IB analysis of anti-PTEN IPs and WCL derived from U2OS cells treated with 30 μM etoposide as indicated time points before harvesting.
(B) Depletion of Nsd2 impaired the di-methylation of Pten. IB analysis of anti-Pten IPs and WCL derived from Nsd2+/+ and Nsd2−/− MEFs treated with 30 μM etoposide for 30 min before harvesting.
(C) K349 was identified as the major di-methylation site on PTEN. IB analysis of anti-HA IPs and WCL derived from 293T cells transfected with the indicated constructs and treated with/without 30 μM etoposide for 30 min before harvesting.
(D) PTEN was detected by the specific K349 di-methylation (K349me2) antibody. IB analysis of anti-HA IPs and WCL derived from U2OS cells transfected with HA-PTEN WT or K349R mutant and treated with/without 30 μM etoposide at indicated time points.
(E) The K349 di-methylation of Pten existed in both cytoplasm and nucleus. IB analysis of anti-Pten IPs from cytoplasm or nucleus as well as WCL derived from NIH3T3 cells treatment with 30 μM etoposide as indicated time points before harvesting.
(F and G) NSD2 deficiency decreased the di-methylation of PTEN at K349. IB analysis of anti-PTEN IPs and WCL derived from U2OS cells stably expressing shNSD2 (F) or Nsd2−/− MEFs (G) that were treated with IR (5 Gy) at indicated time points before harvesting.