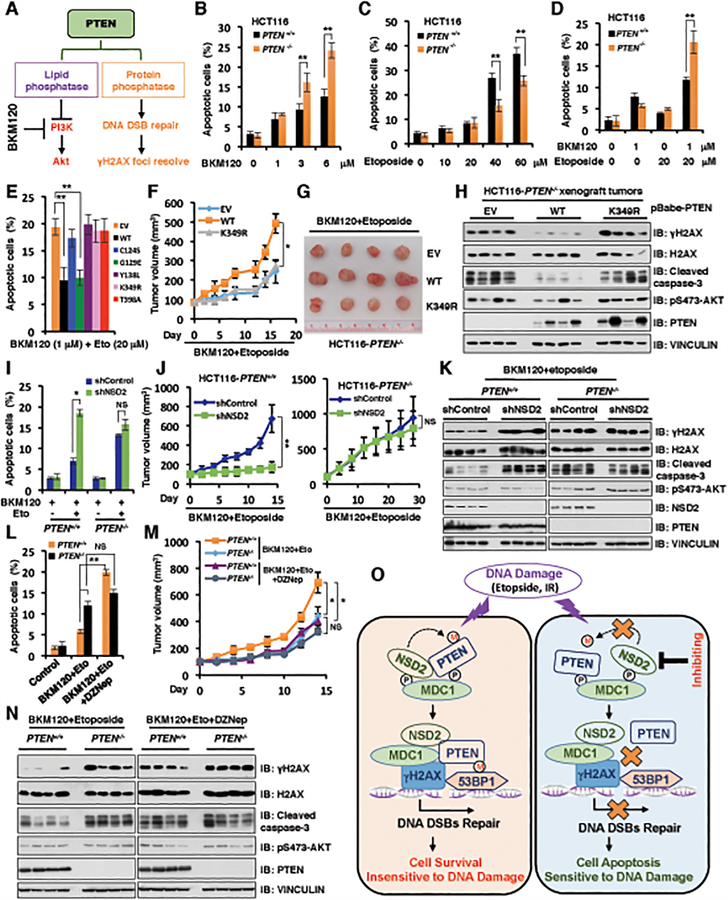
Figure 6. NSD2-mediated methylation of PTEN at K349 dictates cellular sensitivity to DNA-damaging agents.
(A) A schematic model to illustrate that PTEN has lipid phosphatase and protein phosphatase activity, which involves in PI3K/Akt signaling pathway in cytoplasm and DNA DSB repair/γH2AX pathway in nucleus, respectively.
(B and C) PTEN+/+ and PTEN−/− HCT116 cells were treated with increased concentrations of BKM120 (B) for 72 h or etoposide (C) for 48 h, cells were harvested for cell apoptosis assays. Data are represented as mean ± s.d., n = 3, and **p < 0.01 (Student’s t-test).
(D) PTEN+/+ and PTEN−/− HCT116 cells were pre-treated with 1 μM BKM120 for 24 h followed by additional etoposide (20 μM) treatment for 48 h, cells were harvested for cell apoptosis assays. Data are represented as mean ± s.d., n = 3, and **p < 0.01 (Student’s t-test).
(E) PTEN−/− HCT116 cells reconstituted with the indicated PTEN constructs were pre-treated with 1 μM BKM120 for 24 h followed by additional etoposide (20 μM) treatment for 48 h. Cells were harvested for cell apoptosis assays. Data are represented as mean ± s.d., n = 3, and **p < 0.01 (Student’s t-test).
(F-H) Tumor xenograft assays were performed by subcutaneous injection of PTEN−/− HCT116 cells stably expressing PTEN WT, K349R and empty vector (EV). Tumor growth rate in nude mice treated every other day with combination of etoposide (20 mg/kg) and BKM120 (25 mg/kg) was shown (F). Tumors were dissected and recorded after euthanizing the mice (G). IB analysis of the samples was performed using indicated antibodies (H). Four mice each group. *p < 0.05 (Student’s t-test).
(I) PTEN+/+ and PTEN−/− HCT116 cells stably depleting NSD2 by shRNA (with shControl as a negative control) were pre-treated with 1 μM BKM120 for 24 h followed by additional etoposide (20 μM) treatment. 48 h post-treatment, cells were harvested for cell apoptosis assays. Data are represented as mean ± s.d., n = 3, and *p < 0.05 (Student’s t-test).
(J and K) Tumor xenograft assays were performed by subcutaneous injection of PTEN+/+ and PTEN−/− HCT116 cells stably expressing shRNA against NSD2 or shControl as a negative control. Tumor growth rate in nude mice treated every other day with a combination of etoposide (20 mg/kg) and BKM120 (25 mg/kg) was shown in (J). Tumors were dissected after euthanizing the mice and were analyzed by IB with indicated antibodies (K). Statistical analysis of tumor volumes showed significant differences in mean tumor volumes between the shNSD2 and the shcontrol groups. Four mice each group. *p < 0.05, **p < 0.01, NS indicates no significant difference (Student’s t-test).
(L) PTEN+/+ and PTEN−/− HCT116 cells were pre-treated with 1 μM BKM120 for 24 h followed by additional etoposide (20 μM) treatment. 48 h post-treatment, cells were harvested for cell apoptosis assays. Data are represented as mean ± s.d., n = 3. *p < 0.05, **p < 0.01, NS indicates no significant difference (Student’s t-test).
(M and N) Tumor xenograft assays were performed by subcutaneously implanting PTEN+/+ and PTEN−/− HCT116 cells. Tumor growth rate in nude mice treated every other day with a combination of etoposide (20 mg/kg) and BKM120 (25 mg/kg) with DZNep (1 mg/kg) (or with vehicle as a negative control) was shown (M). Tumors were dissected after euthanizing the mice and were analyzed by IB using indicated antibodies (N). Four mice each group. *p < 0.05, NS indicates no significant difference (Student’s t-test).
(O) A schematic model to show the molecular mechanism of NSD2-mediated methylation of PTEN involving in DNA damage repair. DNA damaging agents promote NSD2-mediated di-methylation of PTEN, which is recognized by the 53BP1 tudor domain to recruit PTEN on DNA damage sites and help DNA DSBs repair largely through dephosphorylating γH2AX (left panel). However, inhibiting NSD2 could suppress PTEN di-methylation at K349 and its recruitment on DNA damage sites, which subsequently leads to DSBs repair deficiency and sensitizes cancer cells to DNA damaging agents (right panel). P: phosphorylation; M: methylation.