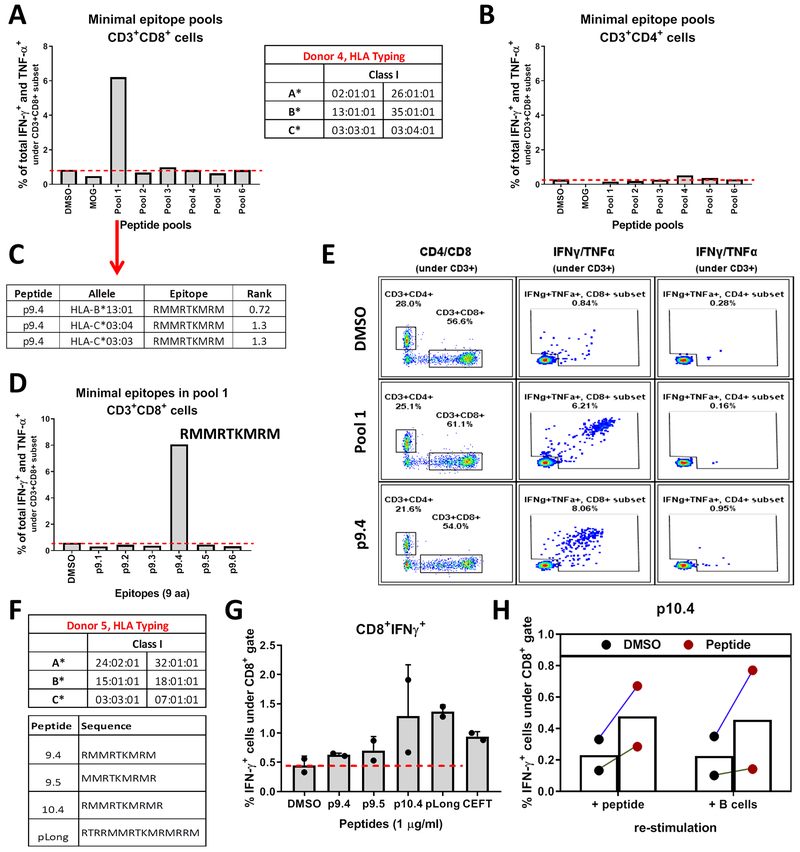
Fig. 6.
T cells recognize mut-CALR epitopes that are endogenously processed and presented. PBMCs from donor 4 were stimulated in vitro with pools of short peptides (9 aa and 6-7 peptides/pool) and frequencies of IFN-γ and TNF-α producing CD8+ (A) and CD4+ (B) T cells were measured by flow cytometry. HLA alleles of individuals were identified by sequence-based genotyping. (C) Binding affinities of donor 4’s alleles to the 6 epitopes within pool 1 were predicted by NetMHCPan 3.0. Epitopes with a predicted percentile rank <2 are listed. (D) Donor 4 PBMCs were stimulated with individual short peptides constituting pool 1. Frequencies of IFN-γ and TNF-α producing CD8+ T cells were measured by flow cytometry. (E) Flow plots showing IFN-γ and TNF-α production by CD8+ and CD4+ T cells upon stimulation with p9.4. (F) PBMCs from donor 5 were stimulated in vitro with peptides listed. (G) Frequencies of IFN-γ producing CD8+ T cells were measured by flow cytometry. Data were pooled from 2 independent experiments. (H) Donor 5 PBMCs expanded with p10.4 were re-stimulated either by the short peptide they were expanded with or by co-culturing autologous B cells that were pulsed with pLong. DMSO or DMSO-pulsed B cells were used as background controls, respectively. Data were pooled from 2 independent experiments.