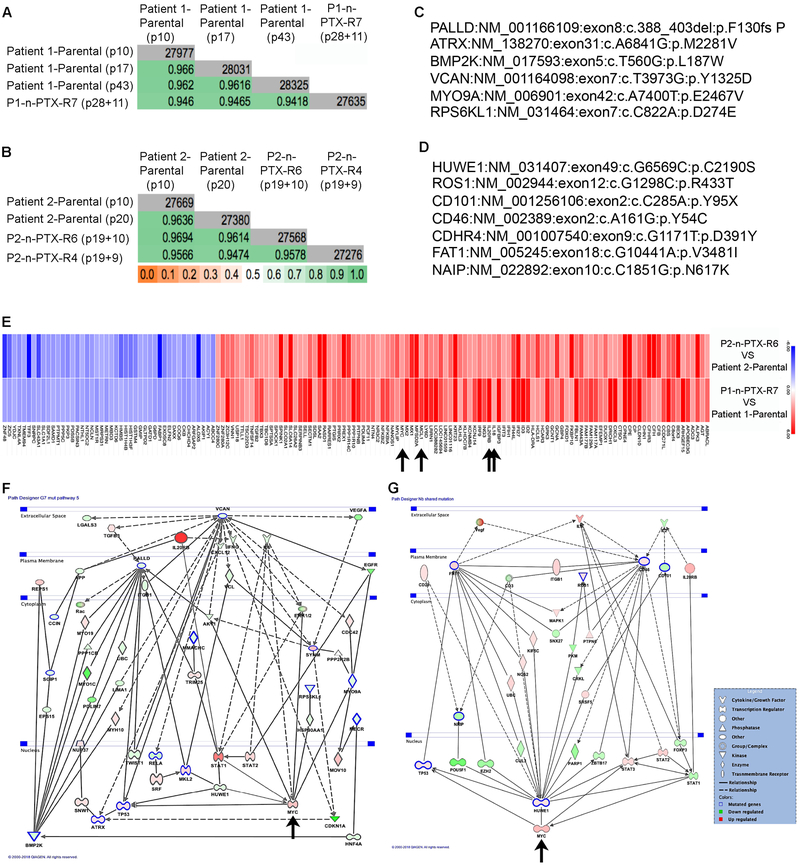
Fig. 5. n-PTX-R cells are genetically stable and have shared mechanisms of nab-paclitaxel resistance.
Jaccard indices (JI) of whole exome sequencing (WES) data of the CR cells from (A) Patient 1 and (B) Patient 2. Gray boxes: total number of genes, green boxes: JI value. The passage numbers at the time of analysis are shown in parentheses. For the n-PTX-R cells, the numbers represent the passage when cell selection began plus the post-selection passage when exome sequencing was performed. (C) Representative non-synonymous mutations in P1-n-PTX-R7 cells vs. Patient 1-parental cells by WES. (D) Shared non-synonymous mutations in P2-n-PTX-R4 (Patient 2-n-PTX-R4) and P2-n-PTX-R6 (Patient 2-n-PTX-R6) cells vs. P2-parental cells by WES. (E). The top 50% shared regulated genes in n-PTX-R cells identified by microarray are shown. Blue: repressed genes, red: induced genes. Arrows: IL20RB, IL18, MCL1 and Myc. Combined mutational and gene expression interactomes for (F) P1-n-PTX-R7 and (G) P2-PTX-R4/6 CR cells. Green shapes: repressed genes in resistant cells, red shapes: induced genes in resistant cells. Blue outlined shapes: mutated genes in the resistant cells, Arrows: Myc.