Fig. 6.
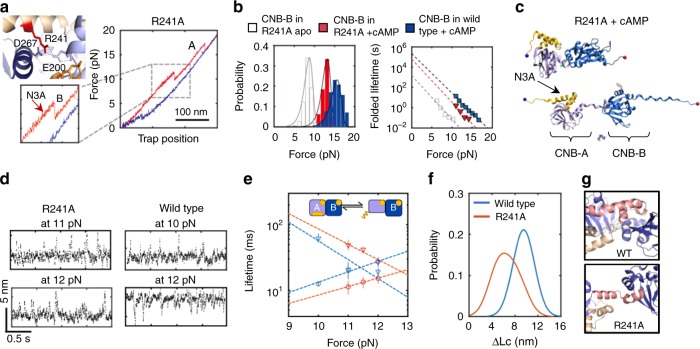
Perturbation of allosteric networks in PKA by mutation R241A. a Residue R241 interacts with both CNB domains through E200 and D267 (PDB 1RGS). Force-extension curve for R241A bound to cAMP (type-III construct S110C/S376C). Zoomed-in is the unfolding rip corresponding to the N3A motif. b Unfolding probability distributions and force-dependent folded-state lifetimes for the CNB-B domain in R241A apo (gray) or bound to cAMP (red). For reference, the wild-type data were included (blue). Solid lines are the unfolding force distribution reconstructed from force-dependent lifetimes. c SMD simulation snapshot of the cAMP-bound R241A protein. d Representative force-clamp trajectories of the N3A motif in R241A and wild type. Source data are provided as a Source Data file. e Force-dependent lifetimes of the N3A motif in the folded (triangles) and unfolded (circles) states for R241A (red) and wild type (blue). Error bars are the SD of different single molecules. f Distribution of the change in contour length (ΔLc) of the N3A motif for wild type (blue) and R241A (red). g The mutation R241A (bottom) hinders interactions established with the CNB-B domain seen in wild type (top). Source data for d and g are provided as a Source Data file