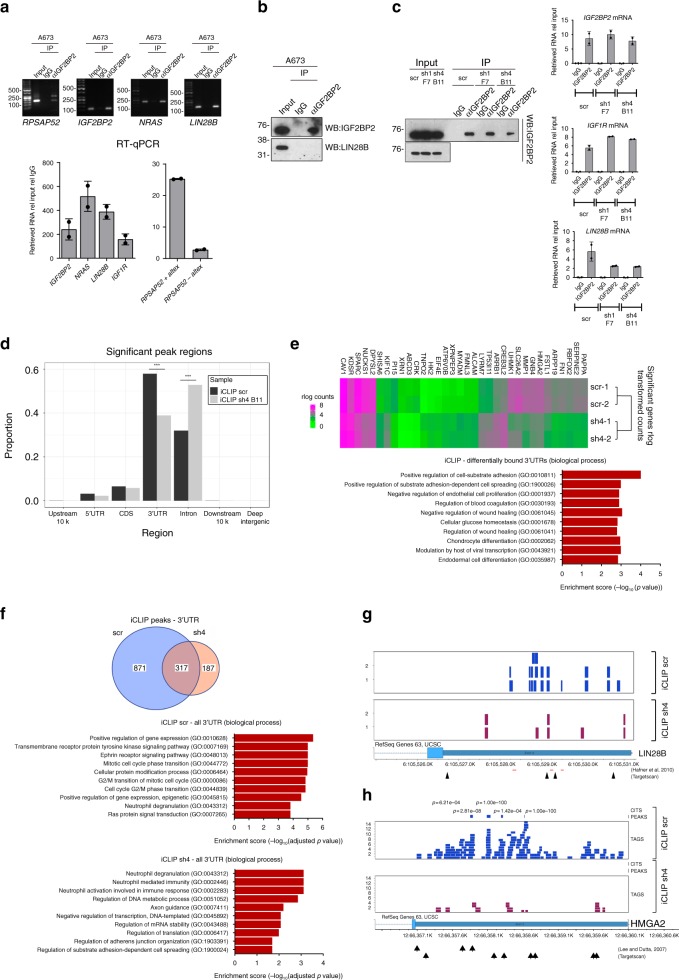
Fig. 5.
Binding of IGF2BP2 to its mRNA targets is affected by RPSAP52 knockdown. a IGF2BP2 was immunoprecipitated from A673 extracts and the pulled-down RNA was analyzed by RT-PCR. The IGF2BP2 and NRAS mRNAs were used as positive controls. Gel images represent semi-quantitative RT-PCR, whereas data on graphs represent means of two independent RT-qPCR analysis ±SD. b Immunoprecipitation of IGF2BP2 from A673 extracts followed by western blot of retrieved proteins. 10% of total extract prior to IP was loaded as control (input). c IGF2BP2 was immunoprecipitated in control or RPSAP52-depleted A673 cells, and the retrieved proteins and RNAs were isolated and analyzed by western blot (left) or RT-qPCR (right), respectively. Graphs correspond to means from two replicates ±SD. d Analysis of IGF2BP2 binding targets from iCLIP-seq experiments in control (scr) or depleted cells (sh4 B11). The absolute number of peaks mapping to 3′UTR regions were 1762 (scr) and 622 (sh4), and to intronic regions were 973 (scr) and 846 (sh4). Asterisks correspond to P-values < 2.2e–16 (two-tailed Fisher’s tests). e Above: heatmap of genes with differential iCLIP counts on their 3′UTR. Results from two experiments are shown. Below: differential enrichment of these genes according to GO biological process categories (top ten are shown). f Above: Venn diagram showing the relation between genes with significant iCLIP peaks present on their 3′UTR regions in the control (scr) or RPSAP52-depleted (sh4) sample. Below: top ten GO enrichment categories (Biological process) for the same genes in the scr or sh4 sample. g UCSC Genome Browser view of LIN28B 3′UTR with the read coverage from IGF2BP2 iCLIP experiment. Previous IGF2BP2-CLIP data positions are shown in red, and predicted let-7 binding sites are indicated by the arrows. h UCSC Genome Browser view of HMGA2 3′UTR with the read coverage from IGF2BP2 iCLIP experiment. Position of significant peaks and CITS are shown above the profiles, and predicted let-7 binding sites are indicated by the arrows. Source data are provided as a Source Data file