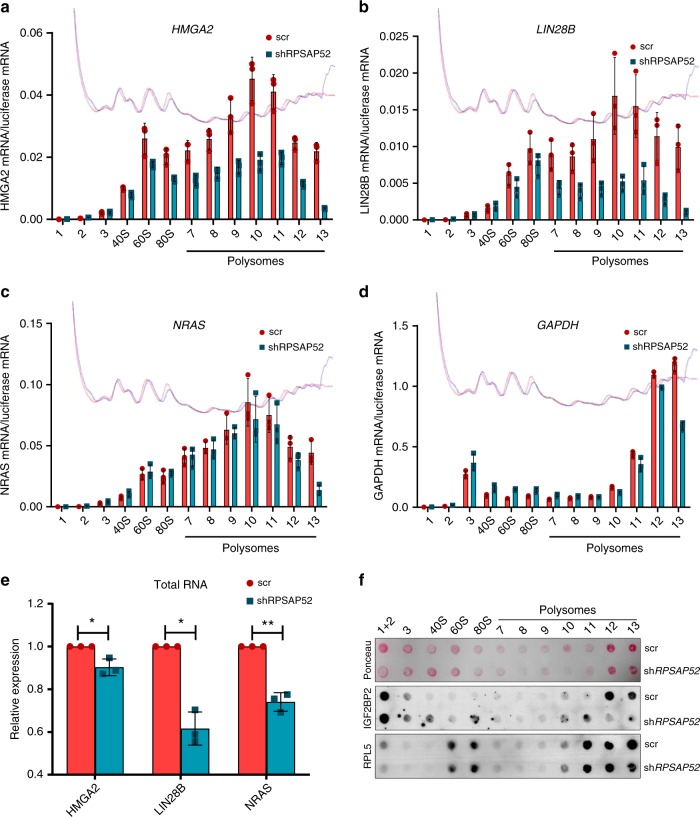
Fig. 6.
IGF2BP2 is redistributed on polysome gradients upon RPSAP52 depletion. a–d Polysome profiles of A673 cells stably depleted of RPSAP52 (shRPSAP52, corresponding to sh4 B11 clone) or control cells (scr). HMGA2 mRNA (a), LIN28B mRNA (b), or NRAS mRNA (c) distribution across the gradient was evaluated in each fraction by RT-qPCR. For comparison, GAPDH mRNA distribution was also assessed (d). Graphs represent the mean ±SD of three replicates. The red and blue lines indicate absorbance at 260 nm for each fraction in control or depleted cells, respectively. e Total RNA from the same cells (n = 3) was analyzed by RT-qPCR. Mean values are shown ±SD. A two-tailed student t-test was used (*P < 0.05, **P < 0.01). f Protein extracted from the 20% of the polysome profile fractions shown in (a–d) were subjected to dot blot analysis with an anti-IGF2BP2 antibody (middle panel) or with anti-RPL5 antibody as control (lower panel). Proteins from 10% of fractions 1 and 2 were loaded together. Membranes were previously stained with Ponceau S (top panel) for loading control. Source data are provided as a Source Data file