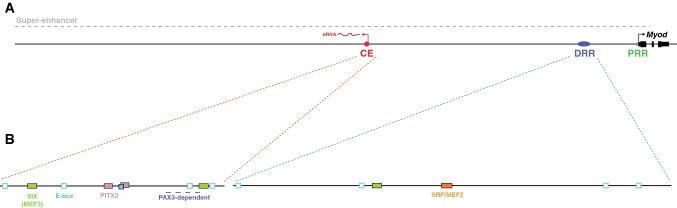
Fig. 1.
Sequences that regulate Myod expression. a Schematic of a region 50 kb upstream of the Myod gene, including the core enhancer (CE, in red), the distal regulatory region (DRR, in blue) and the proximal regulatory region (PRR, in green) immediately upstream of the Myod transcription start site (TSS, black arrow). This region lies within a super enhancer, associated with high levels of transcription factors occupancy, H3K27ac marks and multiple enhancer RNAs (eRNAs). One of these is an eRNA is transcribed from the CE that plays a role in regulating Myod expression through promoting chromatin accessibility at the Myod promoter. b Transcription factor binding sites for bHLH factors (E-box) including MYOD, MYF5, CLOCK, BMAL2, SIM2, MSC and TCF21 are shown in blue, for SIX and EYA factors in green (MEF3 site), for PITX2 in pink (Paired binding site) and for SRF/MEF2 in orange (CaRG box). Closed boxes show sites that have been shown to bind these factors in electromobility shift or other assays are shown as closed boxes, while open boxes show predicted sites. The E-box (closed) that binds CLOCK/BMAL2 overlaps a PIXT2 binding site. A region that is PAX3 dependent in somites and limbs, and is required for expression in myotomally-derived muscles, is indicated by the dashed purple line. (Color figure online)