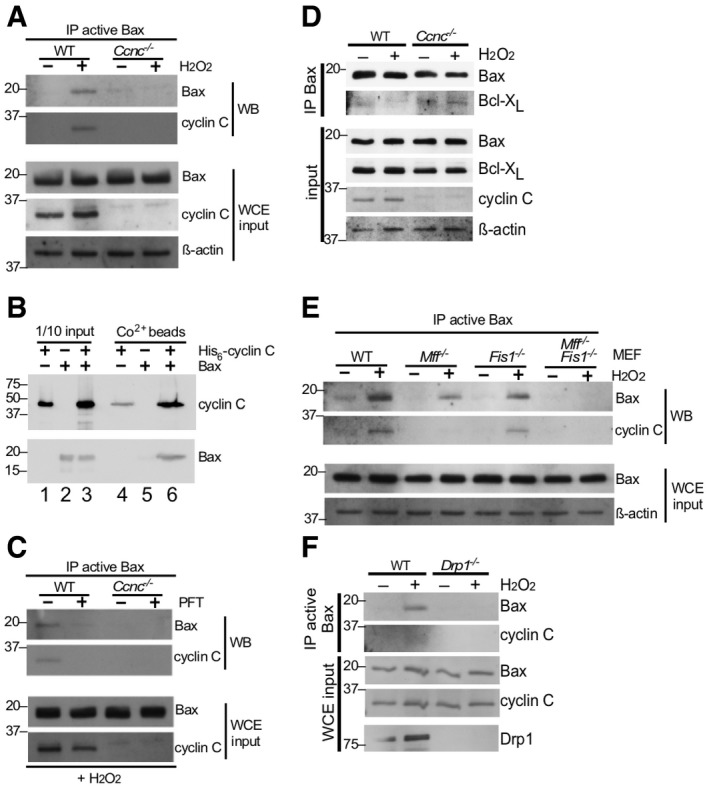
WT and Ccnc
−/− MEF cultures were treated with 0.4 mM H2O2 as indicated and extracts prepared and immunoprecipitated with antibody recognizing the active conformation of Bax. The immunoprecipitates were subjected to Western blot analysis and probed for total Bax or cyclin C. Whole cell extracts (WCE) were subjected to Western blot analysis as indicated to control for protein concentrations in the extracts. Molecular weight markers (kDa) are indicated on the left.
Cyclin C and Bax directly interact. Recombinant Bax and SUMO‐His6‐cyclin C were either incubated together or separately at room temperature and then passed over Co2+ resin. The elutions were subjected to Western blot analysis probing for Bax and cyclin C (lanes 4–6). Input controls for each sample (1/10) are included in lanes 1–3. Molecular weight markers (kDa) are indicated on the left.
Mitochondrial relocalization of cyclin C is required for Bax activation. The experiment described in (A) was repeated except that H2O2‐treated cells were treated with 1 μM pifithrin‐μ (PFT) for 24 h as indicated. Molecular weight markers (kDa) are indicated on the left.
Cyclin C is required for efficient Bax‐Bcl‐XL dissociation. Extracts prepared from MEF cells with the indicated genotypes following treatment with H2O2 (0.4 mM, 4 h) were immunoprecipitated with general Bax antibodies. The immunoprecipitates were subjected to Western blot analysis probing with Bcl‐XL antibodies. Western blot analysis of WCE was performed as input concentration controls. Molecular weight markers (kDa) are indicated on the left.
The mitochondrial fission complex is required for cyclin C‐dependent Bax activation. Extracts were prepared from MEF cultures with the indicated genotypes treated with H2O2 (0.4 mM, 4 h). The extracts were treated as in (A). Molecular weight markers (kDa) are indicated on the left.
Drp1 is required for cyclin C–Bax interaction. Extracts were prepared from wild‐type or Drp1
−/− MEF cultures treated or not with H2O2 and analyzed for the presence of Bax and cyclin C as described in (A). Molecular weight markers (kDa) are indicated on the left.