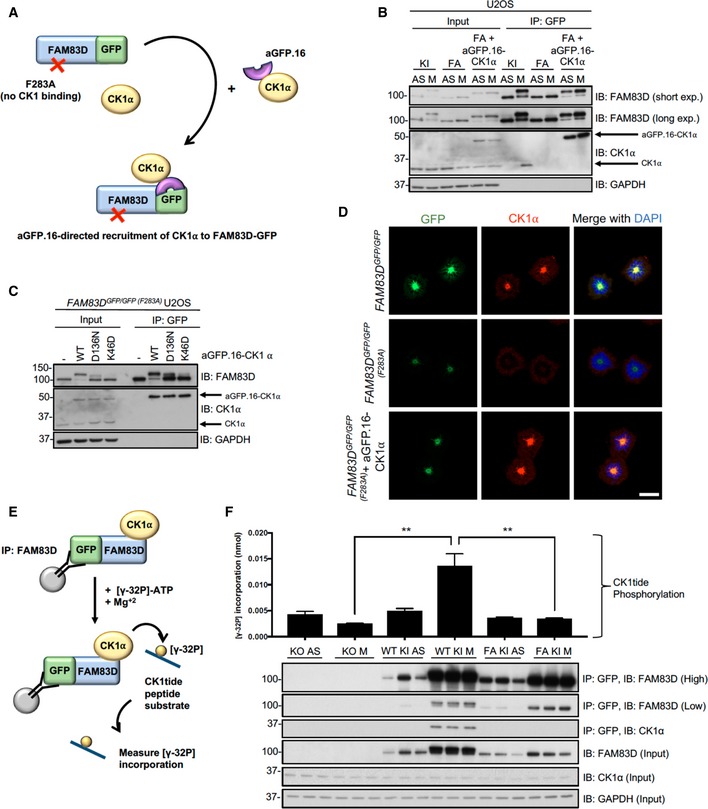
Schematic representation of the anti‐GFP nanobody (aGFP.16)‐based targeting strategy used to deliver CK1α to the CK1‐binding‐deficient FAM83D(F283A)‐GFP mutant.
STLC‐synchronised mitotic FAM83D
GFP/GFP knockin (KI), FAM83D
GFP/GFP (F283A) (FA) and FAM83D
GFP/GFP(F283A) stably expressing aGFP.16‐CK1α (FA + aGFP.16‐CK1α) U2OS cells were subjected to GFP TRAP immunoprecipitation (IP), followed by immunoblotting (IB) with the indicated antibodies. Asynchronous (AS) cells were used as controls.
FA cells were infected with retroviruses encoding wild‐type aGFP.16‐CK1α (WT), or aGFP.16‐CK1α with one of two distinct CK1α kinase‐inactive mutants (D136N or K46D). Uninfected cells (−) were included as a control. Cells were lysed, subjected to anti‐GFP IP and IB with the indicated antibodies.
Immunofluorescence analysis for the cells described in (B) following synchronisation with STLC. Cells were stained using anti‐CK1α antibody, and DNA is stained with DAPI. Scale bars, 20 μm.
Schematic depicting the IP kinase assay strategy used to test whether FAM83D‐bound CK1α was catalytically active.
FAM83D
−/− knockout (KO), KI and FA cells were synchronised in mitosis (M) using STLC. Lysed extracts were subjected to anti‐GFP IPs, followed by [γ32P]‐ATP kinase assays, using an optimised CK1 substrate peptide (CK1tide). AS cells were used as controls. Error bars, SEM; P < 0.01; ANOVA. n = 3. Input and IP samples were analysed by IB with the indicated antibodies.
Data information: All blots are representative of at least three independent experiments.