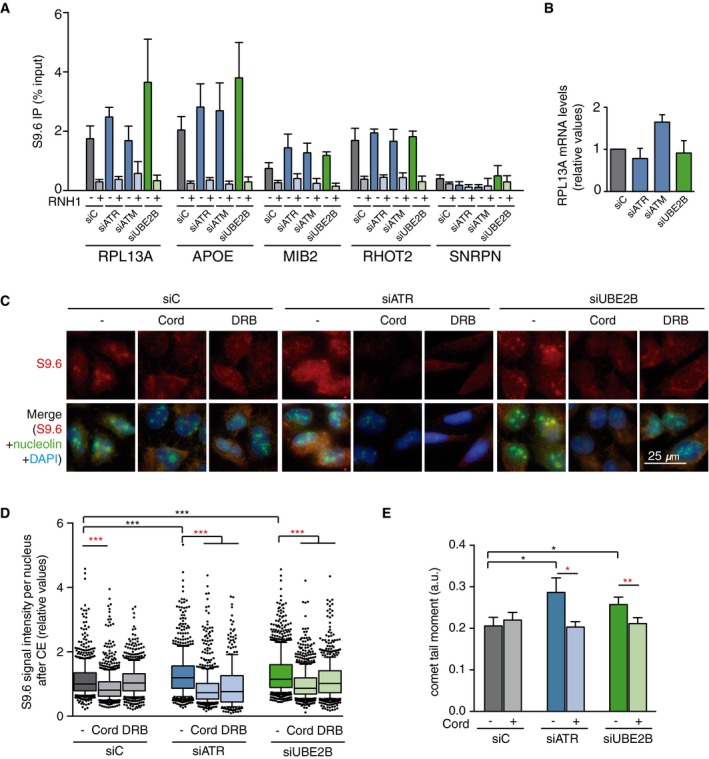
DRIP–qPCR signal values at RPL13A, APOE, MIB2, RHOT2, and SNRPN genes in HeLa cells transfected with the indicated siRNAs and treated in vitro with RNase H pre‐immunoprecipitation where indicated. The mean ± SEM from at least three independent experiments is shown.
Relative mRNA levels from the RPL13A gene in HeLa cells after transfection with the indicated siRNAs. The mean ± SEM from at least two independent experiments is shown.
Representative images of HeLa cells immunostained with S9.6 and nucleolin antibodies after transfection with the indicated siRNAs and after cytoplasm pre‐extraction (CE).
Relative S9.6 signal intensity per nucleus in HeLa cells transfected with the indicated siRNAs and treated with the transcription inhibitors 5,6‐dichloro‐1‐β‐D‐ribofuranosylbenzimidazole (DRB) or cordycepin (Cord). The median of the S9.6 signal intensity per nucleus relative to siC. Boxes and whiskers indicate 25–75 and 10–90 percentiles, respectively. More than 300 total cells from four independent experiments were considered. Values were normalized to the median of siC. ***P < 0.001 (Mann–Whitney U‐test). Black stars denote significant increases, whereas red stars denote significant decreases.
Tail moment from single‐cell alkaline gel electrophoresis (comet assay) of HeLa cells transfected with the indicated siRNAs and treated with the transcription inhibitor cordycepin (Cord). More than 250 total cells were considered. The mean ± SEM of the median from five independent experiments is shown. *P < 0.05, **P < 0.01 (one‐tailed unpaired t‐test). Black stars denote significant increases, whereas red stars denote significant decreases.