Figure 8. CDK12 inhibition decreases transcription elongation rates in bodies of genes with RNAPII processivity defect.
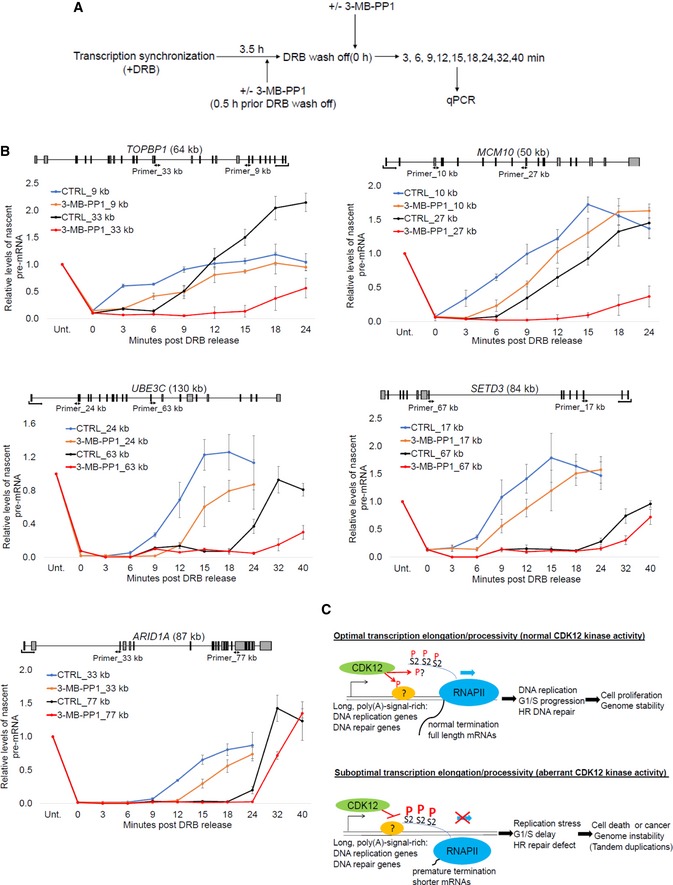
- Experimental outline for measurement of transcription elongation rates. AS CDK12 HCT116 cells were treated with DRB for 3.5 h to synchronize RNAPII at gene promoters. The cells were either pretreated (+) or not (−) with 3‐MB‐PP1 0.5 h prior DRB wash off. After DRB wash off (0 h), fresh medium either supplemented (+) or not (−) with 3‐MB‐PP1 was added and samples were taken at indicated time points for analyses of pre‐mRNA expression by RT–qPCR.
- Transcription elongation rate decreases in bodies of CDK12‐dependent but not CDK12‐independent genes after CDK12 inhibition. Graphs show relative levels of pre‐mRNAs of described genes in AS CDK12 HCT116 cells either treated with 3‐MB‐PP1 or not (CTRL) for indicated times after DRB wash off. Pre‐mRNA levels were normalized to the samples not treated with DRB (Unt) for which the value was set as 1. n = 3 independent experiments, error bars correspond to SEM. Positions of primers (designed to span exon–intron junctions) and their distance from the transcription start site in kb are indicated in the gene structures shown above the graphs.
- Proposed model. Schema shows groups of genes whose RNAPII processivity is particularly sensitive to CDK12 catalytic activity and cellular functions that are especially dependent on optimal expression of these genes. The situation in cells with normal and aberrant CDK12 kinase activity is depicted. CDK12 (green oval) phosphorylates (P) unknown substrate(s) (orange oval), possibly including the CTD (blue line), which results in optimal elongation and processivity (blue arrow) of RNAPII (blue oval) for CDK12‐sensitive genes. Full length, functional mRNAs are synthesized (upper panel). Inhibition of CDK12 leads to hyperphosphorylation (capital P) of Ser2 (S2) in bodies of CDK12‐sensitive genes, which is associated with slower elongation and premature termination. Shorter, aberrant mRNAs are made (lower panel). mRNAs are depicted as black lines.
