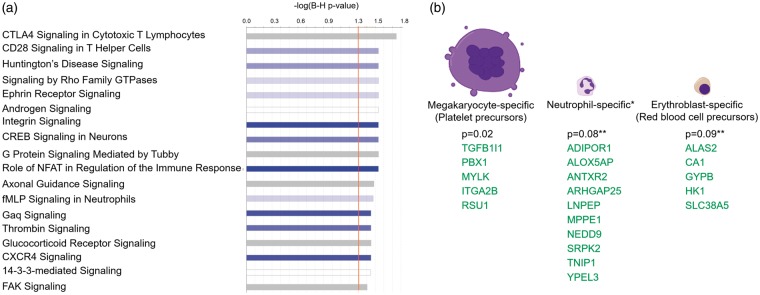
Figure 4.
Over-represented canonical pathways (a) and differentially expressed transcripts from cell type – specific genes (b) in IS versus CTRL. A. The X-axis represents negative log10 (Benjamini-Hochberg corrected p-value). −log10 (Benjamini-Hochberg corrected p-value) > 1.3 corresponds to Benjamini-Hochberg corrected p-value < 0.05. Blue bars – predicted inhibition of the pathways (though no pathway passes Z < − 2 for significant inhibition), grey bars – direction of the pathway cannot be predicted. (b) DET of cell-type specific genes in the 396 DET from IS versus CTRL analysis. P-values represent the hypergeometric probability of overlap between our DET list (by gene symbol) and the list of cell-type specific genes from the HaemAtlas.27 Green – genes with down-regulated DET (expressed lower in IS than in CTRL).