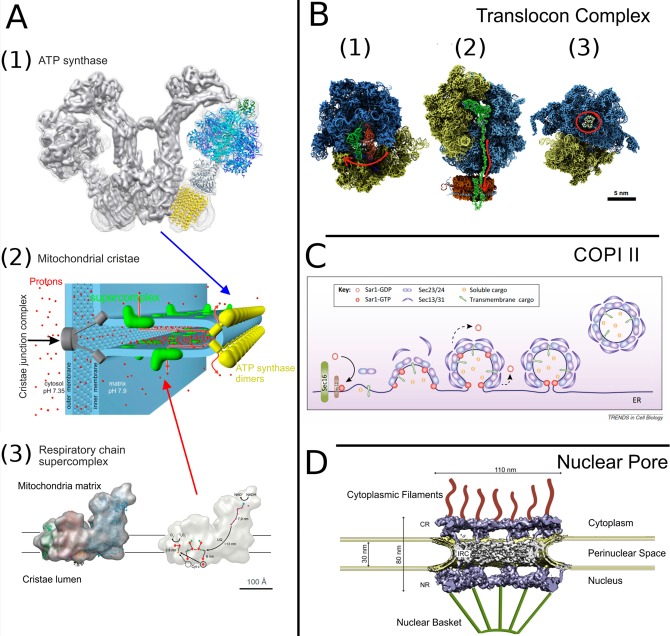
Figure 11.
Membrane-associated protein complexes. (A1) The V-shaped dimer of the mitochondrial ATP synthase. Protomer fitted with atomistic details is taken from PDB 2WSS(338) (α-subunits, cyan; β-subunits, blue; γδε subunits, gray; OSCP, green) and PDB 3U2Y(339) (yellow, c10-ring). Reproduced with permission from ref (333). Copyright 2015 Kühlbrandt. (A2) Organization of mitochondrial cristae. Reproduced with permission from ref (340). Copyright 2015 Davies et al. (A3, left) Cryo-EM structure of the respiratory chain supercomplex composed of NADH dehydrogenase (complex 1, blue), a cytochrome bc1 dimer (complex 3, pink), and cytochrome c oxidase (complex 4, green). (A3, right) The ubiquinol binding sites of complexes 1 and 3. Arrows indicate the electron path through the supercomplex.341 Reproduced with permission from ref (332). Copyright 2011 John Wiley and Sons. (B) Translocon complex. (B1) The pretranslocation state of the ribosome with an A-site tRNA (red) and a P-site tRNA (green). An arrow shows the direction of tRNA’s traversal motion.342 (B2) Insertion of a nascent protein by the ribosome into a nanodisc membrane working with the SecYE translocon. The protein and P-site tRNA are shown in green. An arrow shows the direction of the protein’s movement.343 (B3) Bacterial ribosome with erythromycin (in red circle) at its binding site.344 Reproduced with permission from ref (345). Copyright 2015 Elsevier. (C) Formation of COPII carrier formation. Sar1 is GTPase, Sec23 and Sec24 proteins are the inner-layer coat components, and Sec13 and Sec31 are the outer-layer coat components. Reproduced with permission from ref (346). Copyright 2013 Elsevier. (D) Nuclear pore complex. The tomographic structure (EMD-3105)347 embedded into double membrane of nucleus envelope; cytoplasmic filaments and nuclear basket, which are not part of the experimental structure, are shown in cartoon representation. Reproduced with permission from ref (348). Copyright 2016 Elsevier.