Abstract

Owing to their superior mechanical strength and structure similarity to the extracellular matrix, nanocelluloses as a class of emerging biomaterials have attracted great attention in three-dimensional (3D) bioprinting to fabricate various tissue mimics. Yet, when printing complex geometries, the desired ink performance in terms of shape fidelity and object resolution demands a wide catalogue of tunability on the material property. This paper describes surface engineered biomimetic inks based on cellulose nanofibrils (CNFs) and cross-linkable hemicellulose derivatives for UV-aided extrusion printing, being inspired by the biomimetic aspect of intrinsic affinity of heteropolysaccharides to cellulose in providing the ultrastrong but flexible plant cell wall structure. A facile aqueous-based approach was established for the synthesis of a series of UV cross-linkable galactoglucomannan methacrylates (GGMMAs) with tunable substitution degrees. The rapid gelation window of the formulated inks facilitates the utilization of these wood-based biopolymers as the feeding ink for extrusion-based 3D printing. Most importantly, a wide and tunable spectrum ranging from 2.5 to 22.5 kPa of different hydrogels with different mechanical properties could be achieved by varying the substitution degree in GGMMA and the compositional ratio between GGMMA and CNFs. Used as the seeding matrices in the cultures of human dermal fibroblasts and pancreatic tumor cells, the scaffolds printed with the CNF/GGMMA inks showed great cytocompatibility as well as supported the matrix adhesion and proliferative behaviors of the studied cell lines. As a new family of 3D printing feedstock materials, the CNF/GGMMA ink will broaden the map of bioinks, which potentially meets the requirements for a variety of in vitro cell–matrix and cell–cell interaction studies in the context of tissue engineering, cancer cell research, and high-throughput drug screening.
Keywords: galactoglucomannan methacrylate (GGMMA), cellulose nanofibrils (CNFs), UV cross-linking, mechanical properties, 3D printing, biomedical applications
1. Introduction
Three-dimensional (3D) bioprinting supplies a bottom-up approach patterning soft biocompatible hydrogels to further generate intricately structured scaffolds. These man-made scaffolds allow the understanding of in vivo cell–cell and cell–matrix interactions by integrating the living cells and biomolecules into bioinks and in vitro 3D cell culture.1 The wood-derived nanocellulose family, including cellulose nanofibrils (CNFs) and cellulose nanocrystals produced by top-down methods from wood to elementary nanomaterial, has recently attracted tremendous interests in the field of bioink formulation serving as a platform biomaterial due to its structural similarity mimicking the extracellular matrix (ECM).2−4 Nevertheless, an auxiliary material as a formulation component is often required to tune the ink property. Auxiliary materials such as alginate,5−9 gelatin methacrylate (GelMA),10,11 and other types of cross-linkable polymers4,12 have been studied in improving the printability and shape fidelity of cellulose nanomaterials. Thus, developing novel, versatile, and tunable nanocellulose-based bioinks will advance material and construct design in the implications of bioprinting and other biofabrication techniques.
Inspired by the biomimetic perspective, that is, the integrity of the plant cell wall structure, where the intrinsic affinity of heteropolysaccharides to cellulose provides the cell wall with a composite structure conferring both strength and flexibility,13 hemicelluloses or their derivatives are expected to be promising candidates for a reinforcing cross-linker in nanocellulose-based ink formulation. Hemicelluloses, the second most abundant renewable material after cellulose, are nontoxic, biocompatible, and biodegradable.14,15 The water-soluble and polymeric hemicelluloses could be efficiently and economically fractionated in high purity via modern biorefinery approaches from wood sources.16 Structurally, hemicelluloses present either/both pentose or/and hexose with free hydroxyl groups, which can be easily derivatized such as by tyramine modification,15,17 methacrylate derivatization,18,19 and thiol functionalization,20,21 offering different cross-linking strategies. As a proof of concept, a printable ink formulation based on CNFs and a tyramine-modified xylan has been investigated for printing via enzymatic cross-linking using horseradish peroxidase (HRP) in the presence of H2O2.22 However, the enzymatic cross-linking at slow kinetics might constrain its use in 3D bioprinting, especially when fabricating objects with complex geometry. The UV-induced cross-linking strategy, taking advantage of rapid polymerization in vivo or in vitro, has found numerous applications in engineering tissue equivalents for bone, cartilage, and liver.23−25 The methacrylate group, including the C=C double bond conjugated with the carbonyl group, is a highly useful type of UV-sensitive moieties, easily polymerized by a radical-induced cross-linking. Previously, methacrylated xylan and O-acetyl-galactoglucomannan (GGM) have been prepared in a rough manner involving toxic chemicals that were difficult to remove in the synthesis, such as 4-dimethyl-amino-pyridine (DMAP).17−19 Thus, a greener approach would be needed to synthesize the methacrylated hemicelluloses in the context of bioink formulation. Moreover, the degree of substitution (DS) of methacrylates in the hemicellulose should be well tuned to provide adjustability to the mechanical properties of the engineered scaffold materials.
In tissue engineering, the behavior and function of individual cells are regulated by various directive cues that are spatiotemporally integrated throughout the microenvironment in ECM-mimicking scaffolds.26,27 Tunable biomechanical properties are validated as one of the crucial factors regulating the adherent cell fate.28−31 CNFs were utilized as a flexible 3D model to culture human pluripotent stem cells by Lou et al.32 Later, a 3D printable ink formulation of CNFs and alginate with a high matter loading was investigated along with ionic cross-linking to achieve high stiffness targeting cartilage regeneration using enzymatically and mechanically produced CNFs.6 However, the surface chemistry plays a significant role in the CNFs made by different methods. The CNFs made via 2,2,6,6-tetramethylpiperidine-1-oxyl (TEMPO)-mediated oxidation with a low surface charge were proved to support the improved cell spreading and migration.33 As demonstrated in our recent studies,11,34 the proliferation of fibroblast cells was improved by tuning the mechanical stiffness of the matrix with directly cross-linking CNFs or cross-linking the auxiliary material, referring to GelMA in a binary system of CNF/GelMA. However, the matter loading of TEMPO-oxidized CNFs constrained the tunable material stiffness within a narrow range. Herein, the formulated bioinks based on the TEMPO-oxidized CNFs have limited workability in terms of the tunable mechanical properties, which probably hinders their application potential of engineering a broad spectrum of tissue mimics through bioprinting.
In this work, biomimetic inks were formulated with TEMPO-oxidized CNFs and synthesized UV cross-linkable GGMMAs. An aqueous-based reaction route was established for esterification of the hydroxyl groups on the GGM chains with methacrylic anhydride. Herein, the homogeneous inks of CNFs with GGMMA formed an interpenetrating polymer network (IPN) in the hydrogel. In addition, the structural and biophysical properties of the CNF hydrogels, especially the matrix stiffness and toughness, can be tuned within a wide spectrum. Furthermore, the cytocompatibility of the hydrogel scaffolds printed with CNF/GGMMA inks was preliminarily examined in the cell cultures of human dermal fibroblasts (HDFs) and pancreatic tumor cells (cell line SW-1990). The great cytocompatibility as well as the tunable control over the mechanical stiffness of the hydrogel allow the bioink of TEMPO-oxidized CNF/GGMMA to potentially meet the requirements for a variety of in vitro cell–matrix and cell–cell interaction studies in the context of tissue engineering, cancer cell research, and high-throughput drug screening.
2. Results and Discussion
2.1. Synthesis of GGMMA and Characterization with HPSEC and NMR
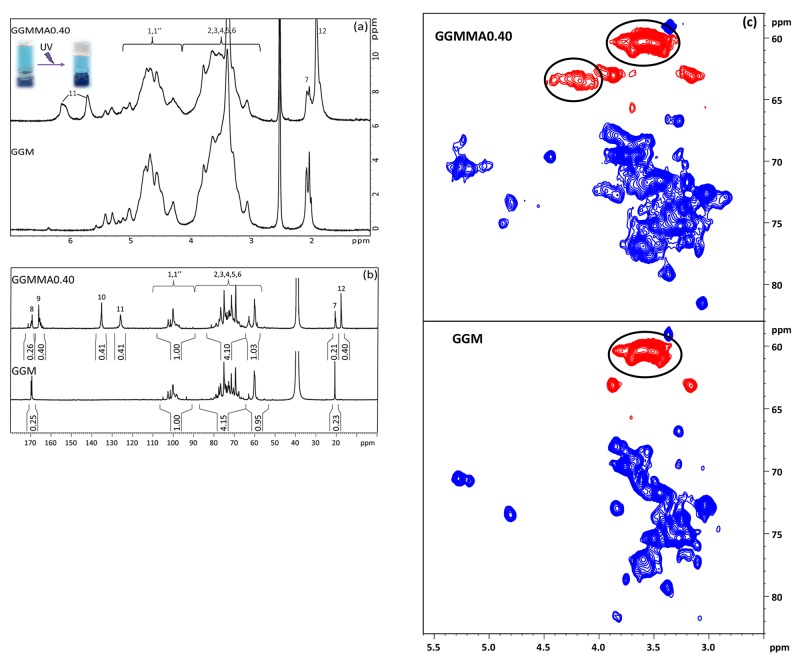
The UV cross-linkable GGMMAs were synthesized by a facile approach for use as an auxiliary component with TEMPO-oxidized CNFs in the ink formulations. In the previous studies,18,19 both methacrylated xylans and GGMMA were synthesized using an organic solvent (DMSO) in the presence of a catalyst of DMAP, which is challenging to be completely removed. A trace existence of DMAP would cause toxicity in the synthesized material. In the present study, the GGMMAs were produced by reacting with methacrylic anhydride in the aqueous-based GGM solution under a mild basic condition at 50 °C without introducing other harmful chemicals. The chemical structures of GGM and GGMMA are illustrated in Scheme 1. According to Figures 1a and S1 (in the Supporting Information), 1H NMR spectra of GGMMA dissolved in DMSO-d6 confirmed the methacrylation of GGM by appearance of the distinctive peaks in the double-bond region (δ 5.5–7.0 ppm) and a sharp singlet that corresponded to the methyl groups (δ 1.9 ppm) in the modified GGM chain. To evaluate the influence of methacrylates on the cross-linking ability of GGMMA, three GGMMAs with varied DS of methacrylation were synthesized. The DS was determined by the quantitative 13C NMR (Figures 1b, and S2, in the Supporting Information). According to the integral comparison based on the anomeric carbon (δ 95–105 ppm), the comparable DS values for each product could be obtained (as shown in Table 1) from the featured peaks of carbonyl carbon (δ 167 ppm), C=C double-bonded carbons (δ 126 and 135 ppm), and methyl carbon (δ 18 ppm). The expectation was that the most frequent and intense esterification reaction is the one taking place at the OH groups located at the C6 carbons on the GGM polymer. Our hypothesis is supported by the multiplicity edited HSQC spectra recorded for the functionalized GGMMA0.40 when compared with the unmodified GGM spectra, as seen in Figure 1b. The esterification of the OH group on the C6 carbon shifts the H6/C6 cross-peaks from the 3.6/60.3 ppm region to the new 4.18/63.4 ppm location, indicated with the black circle in Figure 1c. Some functionalization also occurs at the C2 and C3 hydroxyl groups, but these seem to be less frequent compared to the C6 functionalization. Notably, our structural characterization is consistent with the previous results made by Hannuksela and du Penhoat as well as Ekholm et al.35,36 Additionally, the measured average number molar mass (Mn) of the GGMMAs by HPSEC increased from 9 kDa of GGM to 14 kDa of GGMMA0.25 and GGMMA0.40 due to the increased DS value (Table 1).
Scheme 1. Illustration of the Chemical Structures of GGMMA and GGM.
Figure 1.
(a) 1H NMR spectra of GGMMA0.40 and GGM, (b) quantitative 13C NMR spectra of GGMMA0.40 and GGM, and (c) region of the multiplicity edited HSQC correlation spectra of GGMMA0.40 (top) and GGM (bottom) where red represents negative signal and blue represents positive signal.
Table 1. DS of the Esterified Methacrylate Groups and the Average Molar Mass Distribution of the Unmodified and GGM and GGMMA Materialsc.
DS determined from quantitative 13C NMR.
Mn determined from HPSEC.
Quartz crystal microbalance with dissipation monitoring (DCM-D).
2.2. GGMMA Adsorption by Quartz Crystal Microbalance (QCM) and GGMMA Distribution by Atomic Force Microscopy (AFM) Imaging
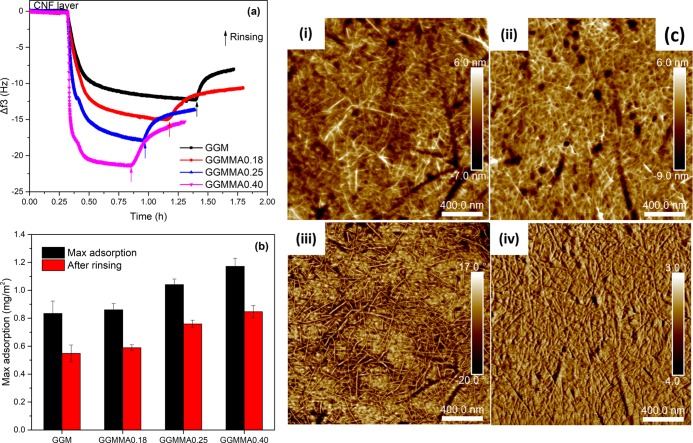
A critical parameter of ink formulation for 3D printing is the good homogeneity of the ink, which refers to the good ink printability without clogging dispensing tips, enabling continuous ink flow to obtain an even strut diameter during extrusion. As demonstrated with QCM-D (Figure 2a), GGM and GGMMAs could irreversibly be adsorbed on the CNF surface. The natural affinity between GGM and cellulose, attributed to the strong hydrogen bonding owing to their similarity of the polymer backbone, is beneficial for achieving the blending compatibility in the ink formulation.14,18,37 The GGMMAs tended to be adsorbed in a greater amount of weight on the CNF surface compared to its native form (Figure 2b), which was revealed by the increase in mass change from 0.55 to 0.8 mg/m2 after the sorption of GGM and GGMMA0.40, respectively. If the mass change was converted into a change in mole amount mmol/m2, the adsorbed amount of GGMMAs was on the same level as that of GGM, indicating that the affinity could be maintained after methacrylation. This is well in line with earlier findings where GGM retained its binding ability to the cellulose surface after functionalization by a hydrophobic moiety of PDMS.38 This was because the GGM-block-PDMS still had a sufficient part of unmodified GGM that has affinity to cellulose, whereas the hydrophobic PDMS block most probably protruded out from the surface. In another study, the adsorption between tyramine-modified xylan and CNFs was weakened due to the introduction of hydrophobicity to xylan derivatives.22 It might be due to the similar structure of the hexose backbones of GGM and CNFs. Moreover, the enhanced adsorption of GGMMA improved the formation of the IPN network and further offered the mechanical strength of the cross-linked hydrogel. Even though the exact amount of binding GGMMA to CNFs within the prepared inks could not be concluded from these modeled measurements, it is believed that the strongly interacted and well-mixed composite inks of GGMMAs and CNFs could boost the tunability of the mechanical properties. AFM images were recorded on the QCM crystals after adsorption to confirm the adsorption of GGMMA on the CNF layer (Figure 2c). Due to the small size of the modified GGMs and the absence of aggregates, no clear difference in topography could be observed after adsorption (Figure 2c(i,ii)). This is well in accordance with previous findings.38,39 The slightly rougher surface observed after GGMMA adsorption, root-mean-square roughness of 1.95 ± 0.28 nm compared to 1.78 ± 0.07 nm for pure CNFs, indicated GMMA adsorption. Surface properties, for instance surface adhesion, and viscoelasticity properties lead to the phase angle change in phase images (Figure 2c(iii,iv)).
Figure 2.
(a) QCM-D adsorption curve of GGMMA onto the CNF layer; (b) adsorbed GGMMA mass on the CNF layer; and (c) AFM images of CNFs in the height mode (i) and phase mode (iii) and of GGMMA0.40 on the CNF layer in the height mode (ii) and phase mode (iv) with a scale bar of 400 nm.
2.3. Ink Formulation and Rheological Property
The viscosities of inks of CNF/GGMMA were studied before printing. Our recent studies demonstrated the printability of TEMPO-oxidized CNFs with a dry matter loading of 1 wt %.11,34 Thus, CNFs prepared with the same approach were adapted for this study. Inks were formulated by mixing GGMMAs of varied DS values with CNFs at different compositional ratios. As shown in Figure 3a, typical shear-thinning profiles were observed for all the ink formulations, which validated the extrudability as indicated also in other studies of printing nanocellulose.11,34 In addition, the decrease of zero-shear viscosity for inks after mixing with GGMMAs also indicated that the entanglement of CNFs was modified after sorption of GGMMA on the nanofibril surface. However, successful 3D printing is dependent not only on the ink extrudability but also on the ability of keeping the printed structure stable in terms of shape fidelity. Several nanocellulose-based ink formulations used for 3D printing employed alginate as an auxiliary material based on the ionic cross-linking with Ca2+, which is reversible depending on the ion concentration.5,6 In addition, ionic cross-linking with divalent ions has also been used to directly cross-link TEMPO-oxidized CNFs owing to the negative charge on the nanofibril surface. However, this is not ideal as it alters the material microenvironment for cells and may need to be removed prior to applications, and the shape resolution tends to decrease when the printed structure is transferred from high to low ionic concentration (such as in phosphate-buffered saline (PBS) solution). Thus, an irreversible covalent cross-linking network via enzymatic cross-linking was investigated by HRP and H2O2.22 However, the required gelation time with a few minutes is quite long. The rate of cross-linking is crucial for biofabrication methods that require gelation during a very short time window.40 The UV cross-linking in the formulation of CNF/GGMMA is based on forming covalent bonds with UV-irradiated radicals and establishing cross-linked networks between GGMMAs in the presence of a photoinitiator. The gelation kinetics was investigated by photorheology. In all the investigated cases, the polymerization was initiated immediately after shedding upon the UV light. The polymerization processes were as short as within 3 min to obtain a leveling-off of the storage modulus (G′). The obtained higher value of G′ indicated that the high degree of cross-linking on GGMMA could create a denser network. The larger the amount of methacrylates in the inks, the longer the UV irradiation time required, which consequently generated a denser GGMMA network within the CNF matrix.
Figure 3.

Viscosity (a) and photorheology (b) profiles of the formulated inks. Note: ink of series I with 1 wt % CNFs and GGMMA0.18 with varying compositional ratios to CNFs of 1:1, 2:1, and 3:1, respectively, referring to IA, IB, and IC; inks IIB and IIC formulated with 1 wt % CNFs as well as 2 and 3 wt % GGMMA0.25, respectively; ink IIIB formulated with 1 wt % of CNFs and 2 wt % GGMMA0.40.
As shown in Figure 3b, ink formulations using the GGMMAs with varied DS values but keeping the same ink loading consistency as well as formulations using the GGMMA with the same DS value but varying the ink loading consistency were investigated. Inks with mixing different DS values and different amounts of GGMMAs exhibited difference in cross-linking time and the maximum G′ value (G′max). Inks of IA, IB, and IC were formulated with GGMMA0.18: the cross-linking rate tended to be higher and the G′max increased largely when more GGMMA0.18 was formulated into the inks. Inks of IB, IIB, and IIIB, which were formulated in the same compositional ratio of 1:2 for CNF/GGMMA but keeping the GGMMAs with increased DS values of 0.18, 0.25, and 0.40, respectively, showed similar trends as when varying the DS values. In comparison with changing DS values, the G′max did not change that much as upon changing the amount of GGMMA. This could be attributed to the small difference in DS values among the GGMMA products. Moreover, the G′max of ink formulation IIC with 3 wt % of GGMMA0.25 and 1 wt % of CNFs reached 60 kPa, which is even higher than that of 17.5 wt % GelMA.41 This indicates that the system of CNF/GGMMA has a great potential to form a strong and lightweight material. By varying the DS and the amounts of GGMMA in the ink formulation, a well-tuned G′ of the biomaterial was displayed after cross-linking, which indicates the tunable strength, mechanical rigidity, and ability to store deformation energy in an elastic manner.13
2.4. Mechanical Property
As the proof-of-concept study, we tested the compressive strengths of the cast discs from the formulated inks. The compressive stress–stain curves of the cast discs are plotted in Figure 4a. The maximum stress under which the disc broke, that is, the yield stress, corresponded well to the measured storage modulus (G′) after UV cross-linking (Figure 3b). The compressive Young’s moduli of the cast hydrogel discs are summarized in Figure 4b. By varying the DS value of GGMMA and the amount of GGMMA in the formulation, the hydrogels’ compressive stiffness can be tuned in the range of 2.5–22.5 kPa. As is well known, the tissue stiffness varies greatly depending on the function of the native tissue in the body.42 Therefore, when engineering tissue equivalents for mimicking the microenvironments of living cells, it is of importance to match the stiffness of the native tissue.26 The broad spectrum of well-tuned stiffness could facilitate the designed ink hydrogels to satisfy the specific applications.43 Thus, the formulated hydrogels with GGMMA and CNFs show a great potential as culture platforms in in vitro cell studies engaging different cell lines as well as highlighting specific physiological phenomena.
Figure 4.

Compressive stress–strain profiles of the cast disc with different ink formulations (a) and their compressive Young’s moduli (b). (Cell image was reproduced with permission from Springer Nature).
2.5. Printability
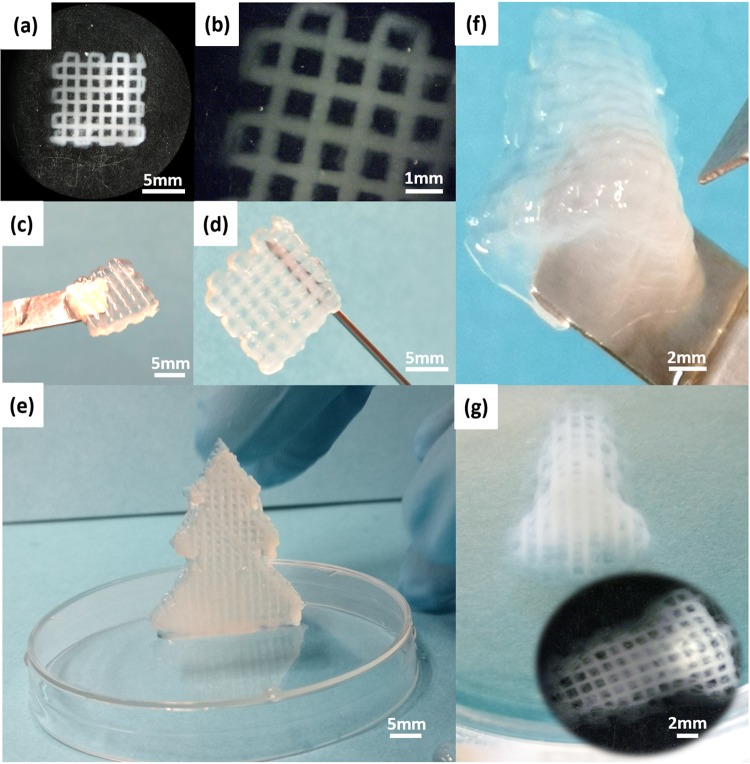
The inks could be dispensed by a printer with a screw-driven printing head and stainless steel tips (27G). The same input flow force and printing speed (5 mm/s) were applied for the inks. The formulated inks with a satisfying viscosity profile (Figure 3) showed good printability as demonstrated in Figure 5. As shown in Figure 5a,b, the printed scaffold kept a well-defined geometry and the printed struts displayed a sharp and distinct edge even at the cross-points. In addition, the printed scaffold was strong enough to keep the structure intact while being lifted with a spatula (Figure 5c) and even with a needle (Figure 5d). The ability of printing big and intricate structures with the formulated inks was demonstrated with the printed scaffold with 4 mm height (Figure S3, in the Supporting Information), the spruce tree model (Figure 5e), and the nose model (Figure 5f,g). Particularly, the printed nose constructs displayed a high resolution of inner structure and good shape fidelity. These features would enhance the performance of these printable inks in the fabrication of objects with complex geometry and hierarchical porosity, aiming at more precisely mimicking the native tissues.44
Figure 5.
Scaffold with high resolution was printed with ink IA after UV cross-linking (a–d). Top view (a), edge view (b), handling with spatula (c), and holding with needle (d) of the printed scaffold with dimension of 10 mm × 10 mm × 2 mm. Standing printed and cross-linked spruce tree model with ink IIB (e). Printed and cross-linked nose model with inner structure in high resolution by ink IA (f) and (g).
2.6. Cytotoxicity, Attachment, and Proliferation of Fibroblasts and Pancreatic Tumor Cells in the CNF/GGMMA Scaffolds
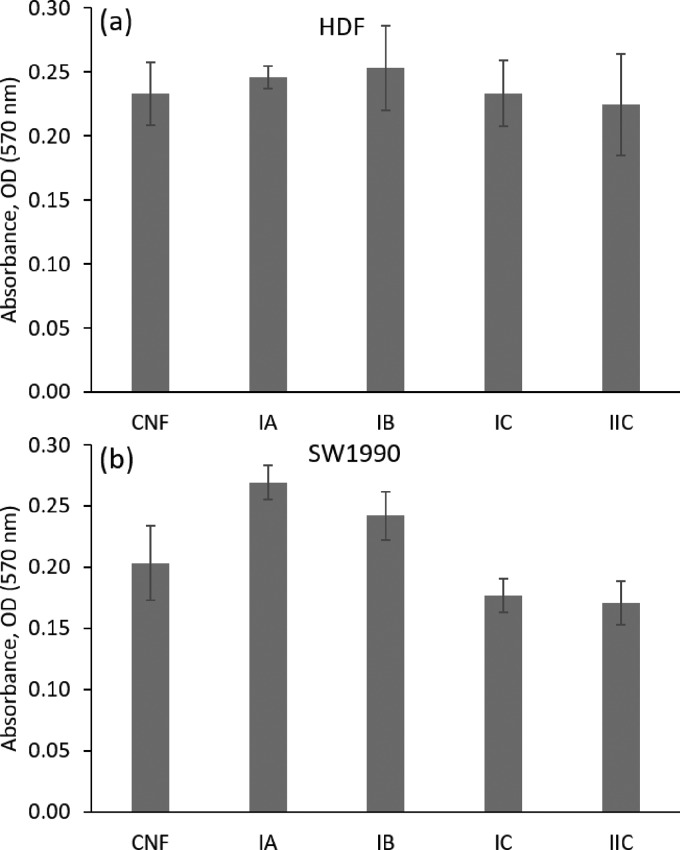
We next assessed the cytocompatibility of the CNF/GGMMA inks, which is the most critical prerequisite for a biomaterial in contact with living cells. The CNFs made via TEMPO-mediated oxidation with a low surface charge were proven to be non-cytotoxic in cell tests of various cell lines, such as fibroblast, HeLa, and Jurkat cancer cells.14,45 In one of our previous studies, CNF hydrogels were reinforced by native GGM and could support the improved cell spreading and migration.14 Here, GGMMA as a facile cross-linker was introduced to tune the mechanical strength of CNF hydrogels; thus, a preliminary study on their cytotoxicity was necessarily performed. The lattice scaffolds (referred to in Figure 5a–d) printed with ink IA, IB, IC, or IIC were studied as the seeding matrices in the culture of a tissue-engineering-relevant cell line HDF and in the culture of a pancreatic tumor cell line SW-1990, separately. After 48 h of cell incubation with the scaffolds, the cell samples were examined according to the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) colorimetric assay to quantitatively measure the capability of viable cells to metabolize the dye 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide. In the culture of HDF, the scaffolds printed with all CNF/GGMMA inks demonstrated satisfactory cell viability compared to the culture on two-dimensional (2D)-Mock (Figure 6a). In the culture of SW-1990 cells, the printed scaffolds showed less comparable viability compared to the culture on 2D-Mock (Figure 6b). The confocal imaging analysis (Figure 6c–h) of the fixed matrices after 48 h incubation also suggests that all the hydrogel matrices supported both types of cells to grow into the 3D matrices, as indicated by the intense distribution of cells positive for two markers of the cytoplasmic marker Phalloidin and the nuclear marker 4′,6-diamidino-2-phenylindole (DAPI). This observation further demonstrates the good cytocompatibility of both investigated cell lines growing in the 3D hydrogel matrices.
Figure 6.
Cell survival rates on the seeding matrices printed with various inks were measured for HDF (a) and for SW-1990 (b) after 48 h of incubation at a density of 5 × 103 cells/96-well by using the MTT assay; bar = mean ± STDEV and n = 3. Representative confocal images of the cells were recorded after 48 of incubation for HDF in the 3D matrix of CNFs (c), ink IB (d), and ink IC (e) and for SW-1990 in the 3D matrix of CNFs (f), ink IB (g), and ink IC (h). The cell morphology is shown by phalloidin (green), and nuclei were counterstained by DAPI (blue). Scale bar: 50 μm.
Next, the cell adhesion into/onto the lattice scaffolds was also quantified after 12 h of cell incubation according to the crystal violet assay, as displayed in Figure 7a for HDF and Figure 7b for SW-1990. The adhesion of HDF onto the hydrogel matrix did not make a significant difference among the tested ink groups (Figure 7a). Compared to the CNF ink, the SW-1990 cells adhered slightly higher onto the hydrogel matrices printed with inks IA and IB; the adhesion onto scaffold matrices printed with inks IC and IIC deteriorated. This is indicative of the attachment of SW-1990 cells on the hydrogel matrices with various degrees of substrate stiffness: hydrogels printed with inks IC and IIC were much stiffer (Young’s modulus situated within 18.5–22 kPa) than the ones printed with inks IA and ink IB (Young’s modulus situated within 5–14 kPa), as indicated in Figure 4b. In native tissues, the majority of cells attach to ECMs of the elastic moduli ranging from 0.01 to 10 kPa.26 It is suspected that the mechanical environment provided by the hydrogel matrices of inks IC and IIC is not favored in the adhesion of the SW-1990 cells.
Figure 7.
Cell adhesion on the seeding matrices printed with various inks was measured for HDF (a) and for SW-1990 (b) after 12 h of incubation at a density of 5 × 103 cells/96-well using the crystal violet assay. Bar = mean ± STDEV and n = 3.
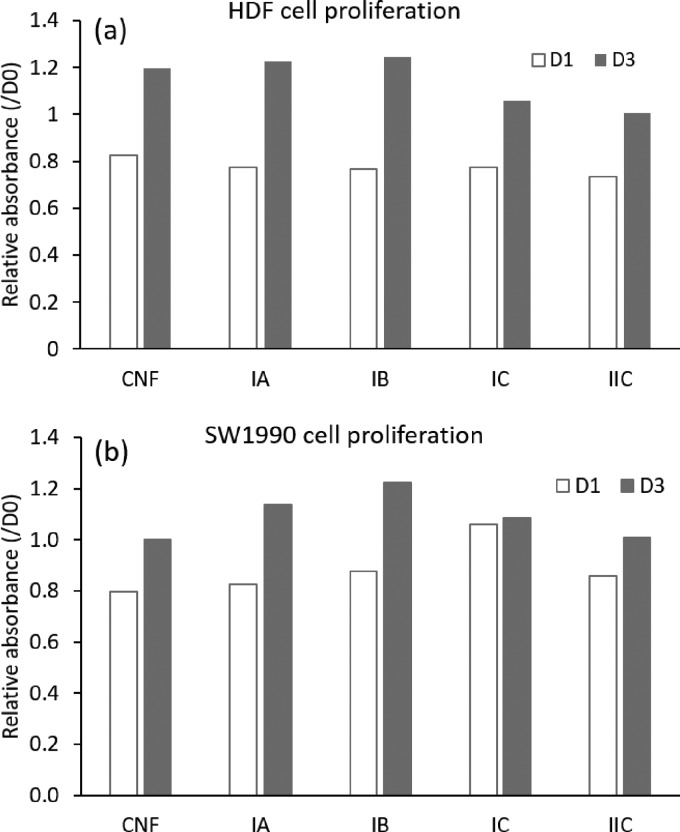
Moreover, the hydrogel scaffolds printed with the CNF/GGMMA inks supported the cell proliferation after longer incubation periods. The cell proliferation was quantitatively evaluated at time points of day 1 (D1) and day 3 (D3) as shown in Figure 8a for HDF and in Figure 8b for SW-1990 cells. At D1, the quantity of HDF cells decreased in all the test groups due to the cell apoptosis. At D3, a higher proliferation rate was observed in hydrogel scaffolds printed with inks IA and IB compared to that in the CNF hydrogel; however, less proliferation was observed in the hydrogel scaffolds printed with inks IC and IIC than in the CNF hydrogel. For the proliferation of SW-1990, a similar proliferative manner was observed at D3 than at D1. Among the test ink groups, the hydrogel scaffolds printed with ink IB supported the proliferation of SW-1990 cells the best.
Figure 8.
Cell proliferation on the seeding matrices printed with various inks was measured for HDF (a) and for SW-1990 (b) after 24 h of incubation (D1) and after 72 h of incubation (D3) at a density of 4 × 103 cells/96-well for HDF and at a density of 3 × 103 cells/96-well for SW/1990 using the MTT assay. The value was reported as the average of two parallel samples.
Rather than focusing on revealing the modulation of matrix mechanical stiffness on the cellular response of a specific cell line, these preliminarily carried cellular assessments were performed to confirm the cytocompability of the developed ink formulations. Compared with other CNF-based bioink systems, for example, CNF/GelMA and CNF/alginate, the CNF/GGMMA ink formulations offer the advantage of tuning the mechanical stiffness of the printed lightweight hydrogel scaffolds in a wide range via facile UV cross-linking (displayed in Figure 4), yet not to mention the favored sustainability of all-wood-originality of the ink components.6,7,11 It is worth stressing that the CNF/GGMMA inks can be formulated to tailor the hydrogel scaffold with well-defined mechanical strength, and the cell-laden bioprinting is of high interests, yet to be further explored with this all-wood-biopolymer formulated bioink toward more specific tissue engineering applications.
3. Conclusions
This study presents a facile and green route to synthesize galactoglucomannan methacrylates with well-controlled DS. Further, the synthesized GGMMAs were blended with 1 wt % TEMPO-oxidized CNFs to formulate 3D printable inks. The intrinsic affinity between these two biopolymers of CNFs and GGMMA modified the rheological properties of the formulated inks. By tuning the DS of GGMMA and the compositional ratio between CNFs and GGMMA, the compressive Young’s moduli of the formulated inks after cross-linking presented a tunable wide spectrum from 2.5 to 22.5 kPa. By the extrusion-based 3D printing technique, scaffolds and intricate objects were successfully printed with high resolution and good shape-fidelity. In summary, the surface modification on CNFs with an easily obtained, efficient, and UV cross-linkable GGMMA is reported here, which offers a reliable strategy for formulating platform biomaterials in the context of tissue engineering. The developed low-concentration ink formulations of CNF/GGMMA present a facile yet effective approach to fabricate lightweight hydrogel scaffolds with a wide spectrum of mechanical properties. Moreover, the as-fabricated hydrogel scaffolds could support the principal cell behaviors including cell viability, adhesion, and proliferation in the preliminary cellular assessments conducted in culture of HDF and SW-1990 cell lines. All-wood-biopolymer formulated inks of CNF/GGMMA are envisioned to broaden the map of bioinks to be used in 3D printing for a variety of in vitro cell–cell and cell–matrix interaction studies.
4. Materials and Methods
CNF dispersion (matter loading of 1.0 wt %, surface charge originated from carboxylic groups of 1.14 ± 0.07 mmol/g) was produced from spruce dissolving pulp (hemicellulose content 4.9%) according to Liu et al.14 The CNFs were processed from softwood pulp by TEMPO/NaClO/NaBr oxidation followed by high-pressure homogenization. GGMs with Mn of 9 kDa were obtained with hot water extraction as previously reported by Xu et al.46 The chemical composition of GGM was analyzed by gas chromatography according to the method mentioned in ref (47) and shown in Table S1. Methacrylic anhydride and Irgacure 2959 were purchased from Sigma-Aldrich.
4.1. Methods
4.1.1. Synthesis of GGMMA
GGMMA was synthesized by reacting GGM with methacrylic anhydride. Briefly, 1 g of GGM was dissolved in 100 mL of deionized water at 50 °C. To this solution, methacrylic anhydride with varying amounts from 1 to 3 mL was added to synthesize GGMMA with varied DS as shown in Table 1. The reactions continued for 3 h at pH 8.0 controlled by adding 5.0 M NaOH. The modified GGMMA was purified by dialysis (cut-off of 2 kDa) for removing unreacted methacrylic anhydride. Purified GGMMA was obtained by lyophilization and stored as dry and protected from light prior to further use.
The synthesized GGMMAs were thoroughly characterized using different spectroscopic NMR techniques and chromatographic HPSEC. All the NMR experiments were performed at 298 K in DMSO-d6 on a Bruker AVANCE III spectrometer operating at 500.13 MHz for 1H and 125.77 MHz for 13C. The quantitative 1H and 13C experiments were recorded using a 5 mm Z-gradient BBO (Broadband Observe) cryoprobe, and the multiplicity edited HSQC (Hetero Nuclear Quantum Coherence, using the pulse program hsqcedetgpsisp2.3) experiments were recorded using a BBO probe. The DS of GGMMA was determined by quantitative 13C NMR analysis recorded with a 10 s pulse delay (D1 = 10 s) and 18 800 scans. The DS was calculated from the integral of the signals given by the esterified methacryl carbonyl carbon (163–167 ppm) and the anomeric carbon of GGM (97–105 ppm). The average molar mass distribution of the original GGM and GGMMA products was determined using HPSEC (Agilent 1100 Series) equipped with a Multi-Angle-Laser-Light-Scattering (MALLS, miniDAWN, Wyatt Technology) detector and an RI (RID, Shimadzu Crop.) detector. The GGM and GGMMA samples with concentration of 2–3 mg/mL were dissolved in water overnight at room temperature. The dissolved samples were purified before any analysis using a syringe filter with a nylon filter (0.2 μm, U.K.). Sodium nitrate with concentration of 0.1 M was used as the eluent for the analysis. During analysis, the flow rate of sodium nitrate was kept at 0.5 mL/min.
4.1.2. Quartz Crystal Microbalance with Dissipation Monitoring (QCM-D )
The adsorption of GGM and GGMMA on CNFs was studied by QCM-D using a Q-Sense E4 instrument (Q-sense, Västra Frölunda, Sweden). A CNF dispersion was prepared by diluting CNF hydrogel with Milli-Q water followed by ultrasonication at 25% amplitude for 1 min using Branson Digital sonifier 450 (Branson Corporation, Danbury, CT). The diluted CNF dispersion was then centrifuged at 8000 rpm for 30 min at 20 °C with an Eppendorf centrifuge 5804R (Eppendorf AG, Hamburg, Germany) to separate CNF fibrils from larger fibril bundles. The supernatant fraction with the finest CNF fibrils was collected for further use.
To enhance retention of CNFs, polyethylene imine with concentration of 0.2 mg/mL was preadsorbed on the surface of the gold-coated quartz crystals. Diluted CNF dispersion was then injected to the QCM-D chamber for absorption of the CNF layer on the crystals. The obtained substrates were thoroughly rinsed with either GGM or GGMMA solution regarding the dry content of CNFs according to the ratio of ink formulation provided in Table 2. The flow rate through the QCM-D chambers was set to 40 μL/min and kept constant during measurements.
Table 2. Ink Formulations with CNFs and GGMMAs.
| GGMMA |
||||
|---|---|---|---|---|
| ink | CNFs (wt %) | type | wt % | compositional ratio between CNFs and GGMMA |
| CNF | 1 | 0 | ||
| IAa | GGMMA0.18 | 1 | 1:1 | |
| IBa | 2 | 1:2 | ||
| ICa | 3 | 1:3 | ||
| IIBb | GGMMA0.25 | 2 | 1:2 | |
| IICb | 3 | 1:3 | ||
| IIIBc | GGMMA0.40 | 2 | 1:2 | |
Ink formulation with GGMMA0.18.
Ink formulation with GGMMA0.25.
Ink formulation with GGMMA0.40.
Simultaneously, changes in frequency and dissipation energy (frictional losses due to viscoelastic properties of the adsorbed layer, ΔD) were monitored. During adsorption, the oscillation frequency of the crystal increases, and deviations from the fundamental frequency (5 MHz) and its overtones (15, 25, 35, 55, and 75 MHz) are detected. According to the Sauerbrey equation (eq 1), the change in frequency (Δf) is proportional to the mass adsorbed per unit surface (Δm).
| 1 |
where C is the sensitivity constant (here C = 0.177 mg/m2) and n is the overtone number (here n = 3). Equation 1 is valid for thin, rigid, and uniform films, but it underestimates mass for viscoelastic films, when ΔD > 1.48 Therefore, the calculated mass values are estimations and should not be considered as absolute values.
4.1.3. Atomic Force Microscopy
The morphologies of CNFs and GGMMA anchored CNFs were studied by Multi Mode 8 atomic force microscopy (AFM) equipped with a NanoScope V controller (Bruker Corporation, Billerica, MA). AFM measurements were done on QCM crystals after adsorption measurements. Tapping mode in air using NCHV-A probes (Bruker) with a tip radius around 10 nm was applied. Flattening was the only correction while analyzing the images with NanoScope 8.15 software (Bruker).
4.1.4. Ink Formulation
Inks for 3D printing were formulated with CNFs and GGMMA with varied DS and compositional ratios. Dried GGMMA powders were mixed into CNF hydrogel by gently shaking, followed by immersing in a water bath at 50 °C. The photoinitiator (Irgacure 2959) with 0.5 w/v % was added for UV cross-linking.
4.1.5. Rheology Tests
The rheological profiles of the formulated inks were studied on a Physica MCR 301 Rheometer (Anton Paar, Austria) with a cone-plate geometry (Ø 50 mm and 1°). Viscosity was recorded with a shear rate from 0.01 to 100 s–1 at room temperature. Oscillation measurements of studying the impact of cross-linking time were conducted with a constant strain and sweep at 1% and 1.5 Hz, respectively. After starting measurement for 2 min, the materials were irradiated upon UV spotlight with an intensity of 10 mW/cm2, and the change in storage modulus was registered.
4.1.6. Mechanical Tests
The compressive strengths of the cast discs from the formulated inks were measured with a Shimadzu EZ-L universal testing machine with a 10 N load cell. The compression speed was set with a constant rate of 0.5 mm/min. The compressive Young’s modulus was calculated according to eq 2
| 2 |
where F is the recorded compressive force, L0 is the original height of the scaffold, A is the area of the scaffold contacted with the upper compression plate, and ΔL was set with a constant displacement of 2 mm.
4.1.7. Printing Tests
Three-dimensional printing of the CNF/GGMMA inks was evaluated with the 3D bioprinter of ROKIT INVIVO (ROKIT, South Korea) equipped with a screw-driven extrusion nozzle. The formulated inks containing 0.5% Irgacure 2959 as the photoinitiator were loaded into a 10 cc syringe. The printing progress was conducted first with layer printing assisted by a drop of 5% Ca2+ solution. After the whole object was printed, post UV cross-linking with a UV light source (Bluepoint4 ecocure, Germany) for 5 min was applied. The wavelength span of the UV source is within UVA radiation of 320–390 nm. The shedding distance is 10 cm from the spotlight, and a UV energy output of about 10 mW/cm2 was expected to deliver to the sample surface, according to the calibration using a radiometer. To check the printability and strut resolution, the inks were tested with the printing of scaffold constructs in the dimensions of 10 mm × 10 mm and height with 2 and 4 mm, respectively. To check the shape fidelity of intricate objects, the spruce tree geometry (30 mm × 30 mm × 4.5 mm) and nose geometry (14 mm × 22.34 mm × 7.88 mm) were printed. All the printing works were conducted with the same parameters including printing speed of 5 mm/s, input flow of 120%, and infill of 15% under the control of Creator K software (South Korea).
4.1.8. Cell Culture
Human dermal fibroblast (HDF) and pancreatic tumor cell line SW-1990 cells were maintained in Dulbecco’s modified eagle medium (DMEM, high glucose) supplemented with 2 mM l-glutamine, 100 IU/mL penicillin and streptomycin, and 10% heat-inactivated fetal bovine serum (FBS). Cells were grown in a 10 cm cell culture plate and kept in a 5% CO2, 37 °C cell culture incubator. The cells were split when the confluency reached 70%.
4.1.9. Cytotoxicity Assay and Proliferation Assay
Cells were washed three times with prewarmed PBS. Trypsin was added into the plate to allow detachment of the cells. Then, cells were collected by using a centrifuge at 1000 rpm for 3 min and resuspended in DMEM for cell number counting. As for the cytotoxicity assay, cells were seeded into a 96-well plate at a density of 5 × 104/mL. After 16 h incubation, different types of ink were introduced into the plate and coincubated with the cells. As for the proliferation assay, HDF cells and SW-1990 cells were seeded into a 96-well plate with densities of 4 × 104/mL and 3 × 104/mL, respectively. After 16 h incubation, different types of inks were introduced into the plates and coincubated with cells for 48 h. Absorbance measurements were carried out using the MTT kit (Sigma) according to the manufacturer’s instruction. The plates were read out at 570 nm of absorbance wavelength using a microplate reader.
4.1.10. Cell Attachment Assay
To quantify the relative numbers of cells maintained on the ink, cell suspensions were kept in contact with the ink for 12 h and nonadherent cells were washed off using warm tap water. The plate was dried by inverting it on the filter paper. Then, 50 μL of 0.5% crystal violet solution was added to each well and incubated at room temperature on a rocker with a frequency of 20 oscillations per minute for 20 min. After three washes using warm tap water, 200 μL of methanol was added into each well and incubated for 20 min at room temperature on a rocker with a frequency of 20 oscillations per minute. The optical density of wells at 570 nm (OD570) was measured with a plate reader.
4.1.11. Confocal Imaging of Cells on the Hydrogel Matrices Printed with the CNF/GGMMA Inks
Samples on coverslips were fixed after 48 h incubation with 4% paraformaldehyde for 30 min and with a mixture of acetone and methanol (1:1) for 5 min on ice. Then, 0.2% Triton X solution in PBS was used to permeabilize the cell membrane. Triton X (10%) in FBS was used for blocking for 2 h at room temperature. The cells were stained with Tom20 primary antibody (Santa Cruz) overnight in a dark, humid chamber. Antimouse Alexa Fluor 488 secondary fluorescent antibody dilution was added the next day and incubated for 30 min. Afterward, the coverslips were counterstained with DAPI blue for 10 min before mounting on glass slides. Confocal images were acquired at room temperature using Zeiss Zen software on a Zeiss LSM880 confocal laser scanning microscope (Carl Zeiss, Inc.) with Plan-Apochromat 10×.
Acknowledgments
W.X. would like to thank the funding from the Johan Gadolin Process Chemistry Centre of Åbo Akademi University. X.W. and C.X. would like to thank the funding from Academy of Finland (298325). O.L. would like to thank the PoDoCo program, the funding received from Jenny and Antti Wihuri foundation, and Mirka Ab for the collaboration. F.C. would like to thank Sigrid Jusélius foundation, the National Natural Science Foundation of China (Grant No. 81702750), and the Basic Research Project of Shenzhen (Grant No. JCY20170818164756460) for funding cell tests. W.X., X.W. and C.X. would like to thank Prof. Gordon G. Wallace and Dr Zhilian Yue at ARC Centre of Excellence for Electromaterials Science, Intelligent Polymer Research Institute, University of Wollongong, Australia, for all the inspiring and fruitful discussions while the work was carried out.
Glossary
Abbreviations
- CNFs
TEMPO-oxidized cellulose nanofibrils
- GGMMA
galactoglucomannan methacrylate
- DS
degree of substitution
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsami.9b03442.
1H NMR and quantitative 13C NMR spectra of GGMMA0.18 and GGMMA0.25 showing the success of GGMMA synthesis; images of scaffolds with different ink formulations indicating the ink printability; chemical composition of the used GGM analyzed by gas chromatography (PDF)
Author Contributions
○ X.Z. and P.Y. contributed equally to this work.
The authors declare no competing financial interest.
Supplementary Material
References
- Murphy S. V.; Atala A. 3D bioprinting of tissues and organs. Nat. Biotechnol. 2014, 32, 773. 10.1038/nbt.2958. [DOI] [PubMed] [Google Scholar]
- Greca L. G.; Lehtonen J.; Tardy B. L.; Guo J.; Rojas O. J. Biofabrication of multifunctional nanocellulosic 3D structures: a facile and customizable route. Mater. Horiz. 2018, 5, 408–415. 10.1039/C7MH01139C. [DOI] [Google Scholar]
- Hoenders D.; Guo J.; Goldmann A. S.; Barner-Kowollik C.; Walther A. Photochemical ligation meets nanocellulose: a versatile platform for self-reporting functional materials. Mater. Horiz. 2018, 5, 560–568. 10.1039/C8MH00241J. [DOI] [Google Scholar]
- Wang B.; Benitez A. J.; Lossada F.; Merindol R.; Walther A. Bioinspired mechanical gradients in cellulose nanofibril/polymer nanopapers. Angew. Chem. 2016, 128, 6070–6074. 10.1002/ange.201511512. [DOI] [PubMed] [Google Scholar]
- Leppiniemi J.; Lahtinen P.; Paajanen A.; Mahlberg R.; Metsä-Kortelainen S.; Pinomaa T.; Pajari H.; Vikholm-Lundin I.; Pursula P.; Hytönen V. P. 3D-Printable Bioactivated Nanocellulose–Alginate Hydrogels. ACS Appl. Mater. Interfaces 2017, 9, 21959–21970. 10.1021/acsami.7b02756. [DOI] [PubMed] [Google Scholar]
- Markstedt K.; Mantas A.; Tournier I.; Martínez Ávila Hc.; Hägg D.; Gatenholm P. 3D bioprinting human chondrocytes with nanocellulose–alginate bioink for cartilage tissue engineering applications. Biomacromolecules 2015, 16, 1489–1496. 10.1021/acs.biomac.5b00188. [DOI] [PubMed] [Google Scholar]
- Heggset E. B.; Strand B. L.; Sundby K. W.; Simon S.; Chinga-Carrasco G.; Syverud K. Viscoelastic properties of nanocellulose based inks for 3D printing and mechanical properties of CNF/alginate biocomposite gels. Cellulose 2019, 581–595. 10.1007/s10570-018-2142-3. [DOI] [Google Scholar]
- Jessop Z. M.; Al-Sabah A.; Gao N.; Kyle S.; Thomas B.; Badiei N.; Hawkins K.; Whitaker I. Printability of pulp derived crystal, fibril and blend nanocellulose-alginate bioinks for extrusion 3D bioprinting. Biofabrication 2019, 10.1088/1758-5090/ab0631. [DOI] [PubMed] [Google Scholar]
- Ojansivu M.; Rashad A.; Ahlinder A.; Massera J.; Mishra A.; Syverud K.; Finne-Wistrand A.; Miettinen S.; Mustafa K. Wood-based nanocellulose and bioactive glass modified gelatin-alginate bioinks for 3D bioprinting of bone cells. Biofabrication 2019, 10.1088/1758-5090/ab0692. [DOI] [PubMed] [Google Scholar]
- Shin S.; Park S.; Park M.; Jeong E.; Na K.; Youn H. J.; Hyun J. Cellulose Nanofibers for the Enhancement of Printability of Low Viscosity Gelatin Derivatives. BioResources 2017, 12, 2941–2954. 10.15376/biores.12.2.2941-2954. [DOI] [Google Scholar]
- Xu W.; Molino B. Z.; Cheng F.; Molino P. J.; Yue Z.; Su D.; Wang X.; Willför S. M.; Xu C.; Wallace G. G. On Low-Concentration Inks formulated by Nanocellulose assisted with Gelatin Methacrylate (GelMA) for 3D Printing towards Wound Healing Application. ACS Appl. Mater. Interfaces 2019, 8838–8848. 10.1021/acsami.8b21268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriksson I.; Gatenholm P.; Hägg D. Increased lipid accumulation and adipogenic gene expression of adipocytes in 3D bioprinted nanocellulose scaffolds. Biofabrication 2017, 9, 015022 10.1088/1758-5090/aa5c1c. [DOI] [PubMed] [Google Scholar]
- Launey M. E.; Ritchie R. O. On the fracture toughness of advanced materials. Adv. Mater. 2009, 21, 2103–2110. 10.1002/adma.200803322. [DOI] [Google Scholar]
- Liu J.; Chinga-Carrasco G.; Cheng F.; Xu W.; Willför S.; Syverud K.; Xu C. Hemicellulose-reinforced nanocellulose hydrogels for wound healing application. Cellulose 2016, 23, 3129–3143. 10.1007/s10570-016-1038-3. [DOI] [Google Scholar]
- Markstedt K.; Xu W.; Liu J.; Xu C.; Gatenholm P. Synthesis of tunable hydrogels based on O-acetyl-galactoglucomannans from spruce. Carbohydr. Polym. 2017, 157, 1349–1357. 10.1016/j.carbpol.2016.11.009. [DOI] [PubMed] [Google Scholar]
- Von Schoultz S.Method for Extracting Biomass. WO Patent WOWO2014009604A1, 2015.
- Kuzmenko V.; Hägg D.; Toriz G.; Gatenholm P. In situ forming spruce xylan-based hydrogel for cell immobilization. Carbohydr. Polym. 2014, 102, 862–868. 10.1016/j.carbpol.2013.10.077. [DOI] [PubMed] [Google Scholar]
- Dax D.; Chávez M. S.; Xu C.; Willför S.; Mendonça R. T.; Sánchez J. Cationic hemicellulose-based hydrogels for arsenic and chromium removal from aqueous solutions. Carbohydr. Polym. 2014, 111, 797–805. 10.1016/j.carbpol.2014.05.045. [DOI] [PubMed] [Google Scholar]
- Peng X.; Ren J.; Zhong L.; Sun R.; Shi W.; Hu B. Glycidyl methacrylate derivatized xylan-rich hemicelluloses: synthesis and characterizations. Cellulose 2012, 19, 1361–1372. 10.1007/s10570-012-9718-0. [DOI] [Google Scholar]
- Maleki L.; Edlund U.; Albertsson A.-C. Thiolated hemicellulose as a versatile platform for one-pot click-type hydrogel synthesis. Biomacromolecules 2015, 16, 667–674. 10.1021/bm5018468. [DOI] [PubMed] [Google Scholar]
- Pahimanolis N.; Kilpeläinen P.; Master E.; Ilvesniemi H.; Seppälä J. Novel thiol-amine-and amino acid functional xylan derivatives synthesized by thiol–ene reaction. Carbohydr. Polym. 2015, 131, 392–398. 10.1016/j.carbpol.2015.06.007. [DOI] [PubMed] [Google Scholar]
- Markstedt K.; Escalante A.; Toriz G.; Gatenholm P. Biomimetic Inks Based on Cellulose Nanofibrils and Cross-Linkable Xylans for 3D Printing. ACS Appl. Mater. Interfaces 2017, 9, 40878–40886. 10.1021/acsami.7b13400. [DOI] [PubMed] [Google Scholar]
- Ifkovits J. L.; Burdick J. A. Photopolymerizable and degradable biomaterials for tissue engineering applications. Tissue Eng. 2007, 13, 2369–2385. 10.1089/ten.2007.0093. [DOI] [PubMed] [Google Scholar]
- Suo H.; Zhang D.; Yin J.; Qian J.; Wu Z. L.; Fu J. Interpenetrating polymer network hydrogels composed of chitosan and photocrosslinkable gelatin with enhanced mechanical properties for tissue engineering. Mater. Sci. Eng. C 2018, 92, 612–620. 10.1016/j.msec.2018.07.016. [DOI] [PubMed] [Google Scholar]
- Yin J.; Yan M.; Wang Y.; Fu J.; Suo H. 3D bioprinting of low-concentration cell-laden gelatin methacrylate (GelMA) bioinks with a two-step cross-linking strategy. ACS Appl. Mater. Interfaces 2018, 10, 6849–6857. 10.1021/acsami.7b16059. [DOI] [PubMed] [Google Scholar]
- Han F.; Zhu C.; Guo Q.; Yang H.; Li B. Cellular modulation by the elasticity of biomaterials. J. Mater. Chem. B 2016, 4, 9–26. 10.1039/C5TB02077H. [DOI] [PubMed] [Google Scholar]
- Paluch E. K.; Nelson C. M.; Biais N.; Fabry B.; Moeller J.; Pruitt B. L.; Wollnik C.; Kudryasheva G.; Rehfeldt F.; Federle W. Mechanotransduction: use the force (s). BMC Biol. 2015, 13, 47. 10.1186/s12915-015-0150-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’brien F. J. Biomaterials & scaffolds for tissue engineering. Mater. Today 2011, 14, 88–95. 10.1016/S1369-7021(11)70058-X. [DOI] [Google Scholar]
- Pelham R. J.; Wang Y.-l. Cell locomotion and focal adhesions are regulated by substrate flexibility. Proc. Natl. Acad. Sci. U.S.A. 1997, 94, 13661–13665. 10.1073/pnas.94.25.13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong H. J.; Liu J.; Riddle K.; Matsumoto T.; Leach K.; Mooney D. J. Non-viral gene delivery regulated by stiffness of cell adhesion substrates. Nat. Mater. 2005, 4, 460. 10.1038/nmat1392. [DOI] [PubMed] [Google Scholar]
- Engler A. J.; Sen S.; Sweeney H. L.; Discher D. E. Matrix elasticity directs stem cell lineage specification. Cell 2006, 126, 677–689. 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- Lou Y.-R.; Kanninen L.; Kuisma T.; Niklander J.; Noon L. A.; Burks D.; Urtti A.; Yliperttula M. The use of nanofibrillar cellulose hydrogel as a flexible three-dimensional model to culture human pluripotent stem cells. Stem Cells Dev. 2014, 23, 380–392. 10.1089/scd.2013.0314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashad A.; Mustafa K.; Heggset E. B.; Syverud K. Cytocompatibility of Wood-Derived Cellulose Nanofibril Hydrogels with Different Surface Chemistry. Biomacromolecules 2017, 18, 1238–1248. 10.1021/acs.biomac.6b01911. [DOI] [PubMed] [Google Scholar]
- Xu C.; Molino B. Z.; Wang X.; Cheng F.; Xu W.; Molino P.; Bacher M.; Su D.; Rosenau T.; Willför S.; Wallace G. 3D printing of nanocellulose hydrogel scaffolds with tunable mechanical strength towards wound healing application. J. Mater. Chem. B 2018, 6, 7066–7075. 10.1039/C8TB01757C. [DOI] [PubMed] [Google Scholar]
- Hannuksela T.; du Penhoat C. H. NMR structural determination of dissolved O-acetylated galactoglucomannan isolated from spruce thermomechanical pulp. Carbohydr. Res. 2004, 339, 301–312. 10.1016/j.carres.2003.10.025. [DOI] [PubMed] [Google Scholar]
- Ekholm F. S.; Ardá A.; Eklund P.; André S.; Gabius H. J.; Jiménez-Barbero J.; Leino R. Studies related to Norway spruce galactoglucomannans: chemical synthesis, conformation analysis, NMR spectroscopic characterization, and molecular recognition of model compounds. Chem. - Eur. J. 2012, 18, 14392–14405. 10.1002/chem.201200510. [DOI] [PubMed] [Google Scholar]
- Eronen P.; Österberg M.; Heikkinen S.; Tenkanen M.; Laine J. Interactions of structurally different hemicelluloses with nanofibrillar cellulose. Carbohydr. Polym. 2011, 86, 1281–1290. 10.1016/j.carbpol.2011.06.031. [DOI] [Google Scholar]
- Lozhechnikova A.; Dax D.; Vartiainen J.; Willför S.; Xu C.; Österberg M. Modification of nanofibrillated cellulose using amphiphilic block-structured galactoglucomannans. Carbohydr. Polym. 2014, 110, 163–172. 10.1016/j.carbpol.2014.03.087. [DOI] [PubMed] [Google Scholar]
- Eronen P.; Junka K.; Laine J.; Österberg M. Interaction between water soluble polysaccharides and native nanofibrillar cellulose thin films. BioResources 2011, 6, 4200–4217. [Google Scholar]
- Ouyang L.; Highley C. B.; Sun W.; Burdick J. A. A Generalizable Strategy for the 3D Bioprinting of Hydrogels from Nonviscous Photo-crosslinkable Inks. Adv. Mater. 2017, 29, 1604983 10.1002/adma.201604983. [DOI] [PubMed] [Google Scholar]
- O’Connell C. D.; Zhang B.; Onofrillo C.; Duchi S.; Blanchard R.; Quigley A.; Bourke J.; Gambhir S.; Kapsa R.; Di Bella C.; Choong P. Tailoring the mechanical properties of gelatin methacryloyl hydrogels through manipulation of the photocrosslinking conditions. Soft Matter 2018, 14, 2142–2151. 10.1039/C7SM02187A. [DOI] [PubMed] [Google Scholar]
- Nemir S.; West J. L. Synthetic materials in the study of cell response to substrate rigidity. Ann. Biomed. Eng. 2010, 38, 2–20. 10.1007/s10439-009-9811-1. [DOI] [PubMed] [Google Scholar]
- Butcher D. T.; Alliston T.; Weaver V. M. A tense situation: forcing tumour progression. Nat. Rev. Cancer 2009, 9, 108. 10.1038/nrc2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyle S.; Jessop Z. M.; Al-Sabah A.; Whitaker I. S. ‘Printability’of Candidate Biomaterials for Extrusion Based 3d Printing: State-of-the-art. Adv. Healthcare Mater. 2017, 1700264 10.1002/adhm.201700264. [DOI] [PubMed] [Google Scholar]
- Liu J.; Cheng F.; Grénman H.; Spoljaric S.; Seppälä J.; Eriksson J. E.; Willför S.; Xu C. Development of nanocellulose scaffolds with tunable structures to support 3D cell culture. Carbohydr. Polym. 2016, 148, 259–271. 10.1016/j.carbpol.2016.04.064. [DOI] [PubMed] [Google Scholar]
- Xu C.; Willfor S.; Sundberg K.; Pettersson C.; Holmbom B. Physico-chemical characterization of spruce galactoglucomannan solutions: stability, surface activity and rheology. Cellul. Chem. Technol. 2007, 41, 51. [Google Scholar]
- Sundheq A.; Sundherg K.; Lillandt C.; Holmhom B. Determination of hemicelluloses and pectins in wood and pulp fibres by acid methanolysis and gas chromatography. Nord. Pulp Pap. Res. J. 1996, 11, 216–219. 10.3183/NPPRJ-1996-11-04-p216-219. [DOI] [Google Scholar]
- Höök F.; Kasemo B.; Nylander T.; Fant C.; Sott K.; Elwing H. Variations in coupled water, viscoelastic properties, and film thickness of a Mefp-1 protein film during adsorption and cross-linking: a quartz crystal microbalance with dissipation monitoring, ellipsometry, and surface plasmon resonance study. Anal. Chem. 2001, 73, 5796–5804. 10.1021/ac0106501. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.