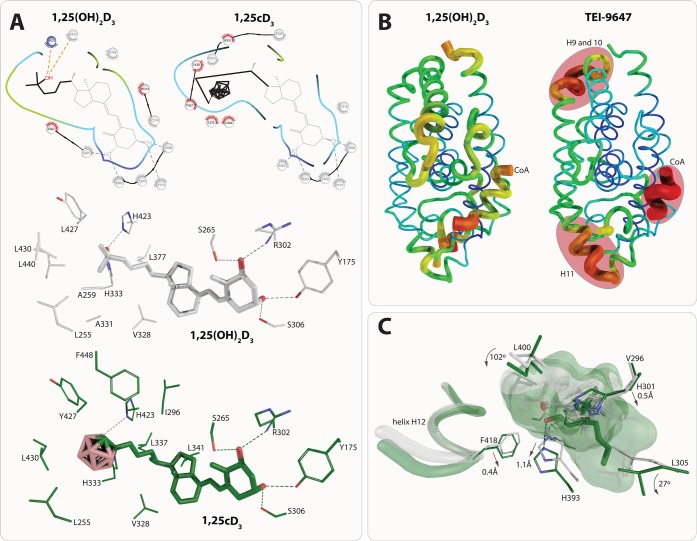
Figure 5.
Structure–function relationship of VDR ligands (I). The carborane group of 1,25cD3 creates additional hydrophobic interactions that compensate for the loss of the 25-OH group. All residues that have conserved interactions are shown in gray (top). Detailed interaction with residues based on PDB code structures 2HC4 (1,25(OH)2D3) and 5E7V (1,25cD3). The displayed interactions are identified under cutoff 3.5 Å (A). Destabilization of the VDR-LBD upon binding of 23,36-lactone analogs. Representation of crystal structure b-factors using structures PDB codes 1RK3 (1,25(OH)2D3, left) and 3A2H (TEI-9647, right). Regions with the highest b-factors are highlighted in red and they are helices H9–10 that may affect heterodimerization with RXR, helix H11 affecting the position of helix H12, and the coactivator peptide showing very high b-factors overall. The most stabile part of the VDR is shown in blue through green, yellow and red monitoring the highest b-factor values (B). Structural implication of 22S-alkyl-2-methylene-19-nor-1,25(OH)2D3 binding. The position of the helix H12 takes the same conformation in both 22S-alkyl-2-methylene-19-nor-1,25(OH)2D3 and 1,25(OH)2D3. Many residues around the two aliphatic chains move or rotate, such as H301 (loop helices H6/7), H393 (helix H11), or F418 (helix H12). Structural elements from 22S-alkyl-2-methylene-19-nor-1,25(OH)2D3 (PDB code 2ZXM) and 1,25(OH)2D3 (PDB code 1RK3) complexes are highlighted in green and white color, respectively (C).