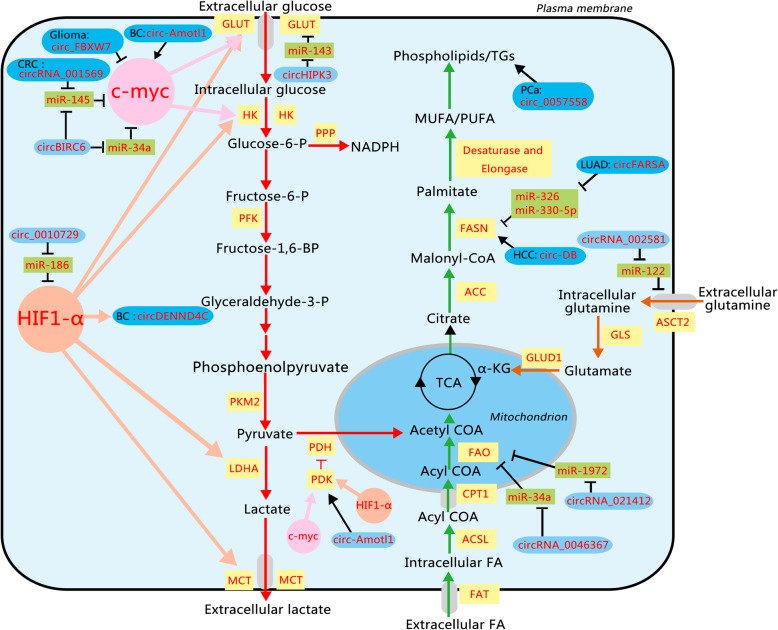
Fig. 1.
Schematic representation of altered metabolic pathways and associated circRNAs in cancer. The characteristic features include two aspects. First, it illustrates deregulated uptake of glucose and amino acids, overactive fatty acid synthesis and oxidation, and NADPH overproduction. Second, it shows how circRNAs interact with these metabolic pathways by enzymes and TFs. HIF1-α and c-myc can adjust the Warburg effect-relevant enzymes and transporters. The majority of circRNAs can antagonize miRNAs to indirectly produce effects. In addition, circ-Amotl1 and circ_FBXW7 can directly affect TFs. The circRNAs in the dark blue rounded rectangle are considered to function in cancer cells, while the circRNAs in the light blue rounded rectangle are just proved to function in normal cells. miRNAs are in the green rectangle; the components affecting metabolism are in faint yellow rectangle. The positive relationships are shown by arrows, and the negative relationships are shown by short dashes. ACC, acetyl-COA carboxylase; ACSL, acyl-CoA synthetases; ASCT2, neutral amino acid transporter2; BC, breast cancer; CPT1, carnitine palmitoyltransferase 1; CRC, colorectal cancer; FAO, fatty acids β-oxidation; FASN, fatty acid synthase; FAT, fatty acid translocase; GLS, glutaminase; GLUD1, glutamate dehydrogenase; GLUT, glutamate dehydrogenase; HCC, hepatocellular carcinoma; HK, hexokinase; LDHA, lactate dehydrogenase A; LUAD, lung adenocarcinoma; MCT. monocarboxylate transporter; PCa, prostate cancer; PDH, pyruvate dehydrogenase; PDK, pyruvate dehydrogenase kinase; PFK, 6-phosphfructa-1-kinase; PKM2, Pyruvate kinase isozymes M2; PPP, pentose phosphate pathway; α-KG, α-ketoglutarate. (Red lines, catabolic pathways; orange lines, glutamine pathways; green lines, lipid pathways)