Abstract
Background
A diagnostic method to simultaneously detect and discriminate porcine circovirus type 1 (PCV1), porcine circovirus type 2 (PCV2) and porcine circovirus type 3 (PCV3) in clinical specimens is imperative for the differential diagnosis and monitoring and control of PCVs in the field.
Methods
Three primer pairs were designed and used to develop a multiplex PCR assay. And 286 samples from 8 farms in Hubei province were tested by the developed multiplex PCR assay to demonstrate the accuracy.
Results
Each of target genes of PCV1, PCV2 and PCV3 was amplified using the designed primers, while no other porcine viruses genes were detected. The limit of detection of the assay was 10 copies/μL of PCV1, PCV2 OR PCV3. The results of the tissue samples detection showed that PCV1, PCV2 and PCV3 are co-circulating in central China. The PCV1, PCV2 and PCV3 singular infection rate was 52.4% (150/286), 61.2% (175/286) and 45.1% (129/286), respectively, while the PCV1 and PCV2 co-infection rate was 11.2% (32/286), the PCV1 and PCV3 co-infection rate was 5.9% (17/286), the PCV2 and PCV3 co-infection rate was 23.4% (67/286), and the PCV1, PCV2 and PCV3 co-infection rate was 1.7% (5/286), respectively, which were 100% consistent with the sequencing method and real-time PCR methods.
Conclusions
The multiplex PCR assay could be used as a differential diagnostic tool for monitoring and control of PCVs in the field. The results also indicate that the PCVs infection and their co-infection are severe in Hubei province, Central China.
Keywords: Porcine circovirus, PCV1, PCV2, PCV3, Viral discrimination, Multiplex PCR
Background
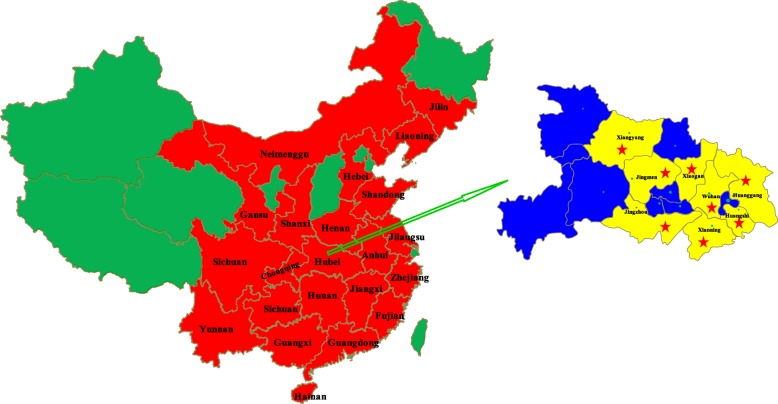
Porcine circoviruses (PCVs), are non-enveloped and circular DNA viruses, which belong to the genus Circovirus, family Circoviridae [1]. At present, PCVs are smallest animal viruses. Two strains of circovirus, PCV1, PCV2, had been proved as infectious to pigs before 2015 [2]. PCV1 was first isolated and in 1974, which just was a contaminant of the PK-15 cell, and nonpathogenic for pigs [3, 4]. However, PCV1 was found that it can replicate efficiently and produce pathology in the lungs of porcine fetuses and have a certain impact on porcine alveolar macrophages [5]. It is a potential damage to the immune system of piglets. PCV2 was first identified from the pigs which was suffering post-weaning multisystemic wasting syndrome (PMWS) in the middle of 1990s [6]. Pigs infected PCV2 have various clinical diseases, which have made the swine industries huge economic losses all over the world [7]. In 2016, a novel circovirus, called PCV type 3 (PCV3), was isolated from diseased pigs in the USA [8, 9]. Subsequently, several outbreaks of it were reported from the United Kingdom [10], Poland [11], Italy, Denmark, Spain [12], Korean [13], Brazil [14], and China [2, 15–17]. PCV1 [5] and PCV3 [2, 8, 12, 13] had been confirmed as potential pathogen associated with many kinds of clinical symptoms, which are similar as PCV2 infection. And now, PCV3 has been found in about 20 provinces in China (Fig. 1).
Fig. 1.
Geographical distribution of PCV3 in China (red regions, till June 2018) and the position of pig farms (red stars) in this study
Both PCV1 and PCV2 infections are common in pig herds all over the world [18], and PCV3 is the third porcine circovirus type found in swine, which is circulating in the swine population [8]. The co-infection of PCV3 with PCV2 was reported in clinical samples of diseased pigs in Hubei province [2]. And co-infection of PCV2 with PCV1 was found in Hubei province [19]. However, PCV1, PCV2 and PCV3 or their co-infections were tested separately using different methods in the previous reports. Considering the high impact of PCV2 and PCV3 on the economy of pig industry, the impact of the potential pathogenic-PCV1, and the similarities between the clinical manifestations associated with PCV3 and PCV2, it is necessary to develop a convenient, sensitive, and specific diagnostic approach to discriminate PCV1, PCV2 and PCV3 infection.
However, there is no convenient and specific diagnostic assay capable of differentiating PCV1, PCV2 and PCV3 infection. Therefore, in present study, a, simple, specific and sensitive multiplex PCR assay was developed to detect and discriminate PCV1, PCV2 and PCV3 in clinical specimens. The accuracy and applicability of the multiplex PCR were evaluated for detection of PCVs DNA in clinical samples collected from the eight pig farms in Hubei province (Fig. 1) where co-infection of PCVs was reported [2].
Methods
Cells and viruses
PK15 cells were cultured in DMEM supplemented with 10% fetal bovine serum (Gibco) at 37 °C in an incubator with humidified 5% CO2. PCV1, PCV2, PCV3 and other viruses, including Torque teno sus viruse type 1 (TTSuV1), Torque teno sus viruse type 2 (TTSuV2), pseudorabies virus (PRV), porcine parvovirus (PPV), rotavirus (RV), Japanese encephalitis virus (JEV), porcine epidemic diarrhea virus (PEDV), porcine deltacorona virus (PDCoV), stored at Key Laboratory of Prevention and Control Agents for Animal Bacteriosis (Ministry of Agriculture), were used to verify the specificity of the developed multiplex PCR assay. And the viruses PCV1, PCV2, PCV3 were also used as positive control viruses. Viral genomic DNA/RNA for the specificity of the proposed multiplex PCR assay were extracted according to our previous study [20].
Primers design
The multiple sequences in GenBank for each virus were aligned including PCV-1 (n = 12), PCV-2 (n = 12), and PCV-3 (n = 12) that covers 85.71, 1.07, and 5.36% of the sequences in GenBank for the respective virus. The nucleotide sequences were submitted to BLAST using MEGA 5.10 software. Three pairs of primers were designed based on the characteristic of the PCV strains (Table 1). The PCR-amplified PCV1, PCV2 and PCV3 products were 310, 505 and 1021 bp, respectively. The primers were synthesized by Sangon Biotech Co., Ltd. (Shanghai, China).
Table 1.
Sequences of the primers for multiplex PCR
| Primer | Primer sequences (5’-3’) | Origin/target gene | Location | Products |
|---|---|---|---|---|
| PCV1-F | GAAAGTGAGCGGGAAGAT | PCV1 (GenBank: KX827790.1) /Rep | 499–516 | 310 bp |
| PCV1-R | CTGATTGCTGGTAATCAA | 790–808 | ||
| PCV2-F | CACATCGAGAAAGCGAAAGGAAC | PCV2(GenBank: MG229682.1) /Rep | 294–316 | 505 bp |
| PCV2-R | TGCGGGCCAAAAAAGGTACAGTT | 776–798 | ||
| PCV3-F | AGCAGTGCTCCCCATTGA | PCV3(GenBank: KX898030.1) /Rep, Cap | 1431–1448 | 1021 bp |
| PCV3-R | TGGGCCCGACCAAATCCGG | 428–446 |
DNA/RNA extraction
Viral DNA/RNA samples for proposed multiplex PCR were extracted using the Viral DNA/RNA Miniprep Kit (Axygen Scientific, CA, USA) from the tissue samples. Total DNA/RNA was eluted with 30 μL of diethyl pyrocarbonate-treated water, used immediately or stored at −80 °C. cDNA was constructed by M-MLV reverse transcriptase (Promega, Madison, WI, USA) with 5–7 μL RNA (RV, JEV, PEDV and PDCoV) .
Single PCR and the standard templates construction
The PCR was performed in a 50 μL reaction mixture including 5.0 μL 10 × PCR buffer, 4 μL dNTPs (10 mM of each), 1.0 μL primer (10 μM), 2.5 U Taq DNA polymerase (5 U/μL) (TaKaRa, Dalian, China), 3 μL DNA (100 copies/μL), and added distilled water to 50 μL.
The amplifications were performed under the following conditions: after initial denaturation at 95 °C for 4 min, 35 cycles were conducted at 94 °C for 30 s, 55 °C for 40 s and 72 °C for 60 s, followed by a final extension at 72 °C for 10 min. Each specific viral target fragment was cloned into the plasmid pMD18-T (TaKaRa). The constructed recombinant plasmids were sequenced and confirmed to use as standard templates for optimization of the following PCR assays.
Optimization of the multiplex PCR
In order to obtain the best reaction parameters, the multiplex PCR was optimized by varying single parameters while other parameters were maintained.The optimization was performed in a 50 μL PCR reaction mixture as follows: 10 × PCR buffer 5.0 μL, 10 mM dNTPs 2–4 μL, each 10 μM primer (Table 1) 0.5–1 μL, Taq DNA polymerase (5 U/μL) (TaKaRa, Dalian, China) 0.5–1 μL, the DNA template 3.0–5.0 μL(100 copies/μL), and added distilled water to 50 μL.
The amplifications were performed under the following conditions in a thermal cycler (Bio-Rad, Hercules, CA, USA). After 5 min initial denaturation at95 °C, 35 cycles were conducted at 94 °C for 40 s, 52–58 °C for 40 s and 72 °C for 50–70 s, followed by a 10-min final extension at 72 °C. The PCR products were detected according to our previous study [20].The specific viral target fragments were clonedinto the plasmid pMD18-T (TaKaRa, Dalian, China).
Plasmids containing the PCV1, PCV2 or PCV3 gene were purified using a MiniBEST Plasmid Purification Kit (TaKaRa, Dalian, China). Each plasmid sample concentration was determined by measuring the absorbance at 260 nm using a Eppendorf BioSpectrometer (Eppendorf, Hamburg, Germany), and each cloned gene copy number was quantified as previously described [21]. The standard PCV1, PCV2 and PCV3 DNA samples were ten-fold diluted (from 107 to 10−2copies/μL) and stored at − 80 °C until use.
Specificity of the multiplex PCR
Specificity of the multiplex PCR assay was determined by using the DNA or cDNA of above-mentioned porcine viruses as templates and ddH2O as a negative control. All templates and ddH2O were repeated three times for specificity of the multiplex PCR. PCV1, PCV2 and PCV3 were verified by sequencing and the other virus strains (TTSuV1, TTSuV2, PRV, PPV, RV, JEV, PEDV and PDCoV) were identified by serological or PCR methods.
Sensitivity of the multiplex PCR
Total DNA from the plasmids was extracted and used to detect the sensitivity of the developed multiplex PCR assay. The ten-fold diluted standard PCV1, PCV2 or PCV3 DNA samples (from 107 to 10−2copies/μL) were used as templates for the multiplex PCR. All samples were repeated in triplicate for sensitivity of the multiplex PCR.
Interference test of the multiplex PCR
To investigate whether the different viral concentrations could influence the detection efficiency of this multiplex PCR assay, 10-fold serial dilutions (10–1–102 copies/μL) in distilled water were prepared to extract the PCVs DNA. Two combinations were tested: (i) the PCR mixture containing three primer pairs and three templates containing equal volumes of DNA from different dilutions, and (ii) three primer pairs and three templates containing equal volumes of DNA from same dilutions.
Clinical sample collection
A total of 286 tissue samples including lung, spleen and lymph node, were collected from diseased pigs from eight farms in Hubei Province (Fig. 1), central China, from October, 2017 to March, 2018. They are commercial source. They were collected and sent to us by the owners to detect PCVs. These pigs were suspected to have clinical signs of PCVs. Specifically, the clinical signs of the diseased pig were as follows: 92 samples of reproductive failure cases, 78 samples of pigs with PMWS, 65 samples of respiratory disorders cases, 36 diarrhea samples and 15 PDNS samples. All samples were collected in accordance with the standards for animal welfare approved by the Animal Ethics Committee of the Hubei province.
Sample preparation
The tissue samples were cut into pieces in a homogenizer, added five times quality of phosphate-buffered saline (PBS, 0.1 M, pH 7.2) and homogenized. All samples were frozen and thawed three times, and centrifuged at 12,000 rpm for 5 min at 4 °C. The supernatants were used for DNA extraction immediately or stored at − 80 °C until use.
Detection of PCVs
The DNA of the clinical specimens were detected using the multiplex PCR assay to investigate the epidemiology of PCVs in Hubei province, central China. To confirm the accuracy of the developed protocol, each PCR product of positive samples was cloned into the plasmid pMD18-T and sequenced. And two real-time PCR methods [22, 23] were used to detect PCV1, PCV2 and PCV3 for comparison.
Animal experiment
Twenty PCV free pigs were average divided into four groups randomly, PCV1, PCV2 and PCV3 experiment group and control group. The pigs are experimental source. They were derived from the farm of Institute of Animal Husbandry and Veterinary, Hubei Academy of Agricultural Sciences. Each pig of the experiment group was injected with 1 mL PCV1, PCV2 or PCV3 inoculums (>106.0TCID50/mL) intravenously, respectively. The control group was injected with 1 mL of sterile physiological saline. Each group was fed separately. Two weeks later, all the pigs were euthanized (CO2 inhalation) to collected tissue samples. Viral DNA was extracted as described above and used as templates to detect PCV1, PCV2 or PCV3 using the established multiplex PCR.
Results
Optimization of the multiplex PCR assay
The optimum parameters of the proposed multiplex PCR were as follows. A final 50-μL volume of master mix for the multiplex PCR including 10 × Buffer 5.0 μL, Taq polymerase (5 U/μL) 0.5 μL, DNA template 5.0 μL, dNTPs (10 mM) 4 μL, each primer (10 μM) 1.0 μL, and nuclease-free water was added to make a total volume of 50 uL per reaction. An optimized experimental protocol consisted of a 5-min denaturation program at 95 °C, and 35 cycles amplification program (denaturation at 94 °C for 40 s, 56 °C for 40 s, and elongation at 72 °C for 50 s), followed by an extension at 72 °C for 10 min.
DNAs of PCV1, PCV2, PCV3, and ddH2O, the negative control, were detected using the protocol described above and the multiplex PCR amplification results are illustrated in Fig. 2.
Fig. 2.
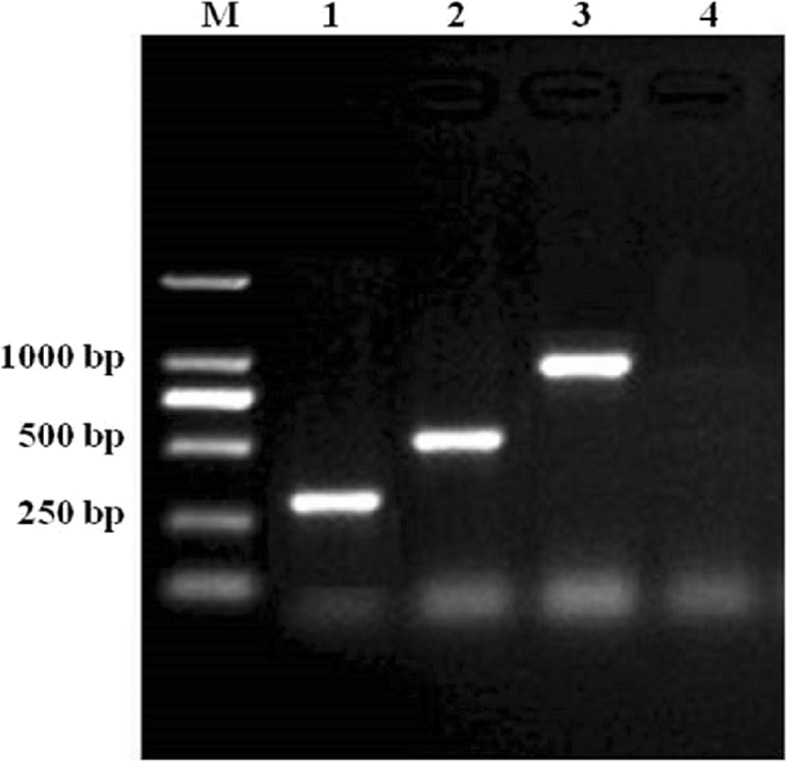
Electrophoresis of multiplex PCR products in optimization conditions. Lane M, DL2000 DNA marker; lane 1, PCV1; lane 2, PCV2; lane 3, PCV3; lane 4, ddH2O
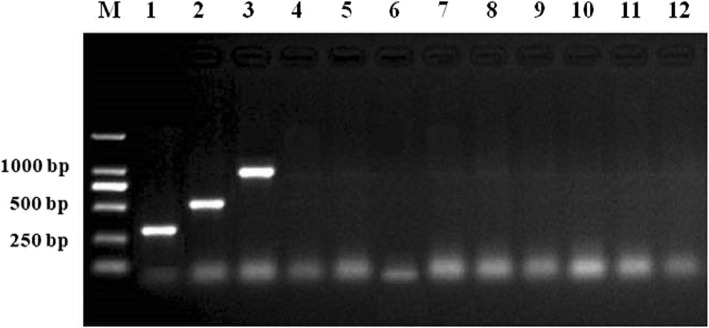
Specificity of the proposed multiplex PCR
The specificity of the three primer pairs for PCVs was analyzed using the developed multiplex PCR. Each DNA/cDNA of the virus mentioned above was amplified using the three defined primer pairs in a reaction respectively. As illustrated in Fig. 3, the multiplex PCR assay was specific for PCVs because no amplification products occurred with TTSuV1, TTSuV2, PRV, PPV, RV, JEV, PEDV, PDCoV and ddH2O (lanes 4–12), whereas the PCV1, PCV2 and PCV3 target genes were specifically amplified using the three defined primer pairs (lanes 1–3).
Fig. 3.
Specificity of multiplex PCR for the detection of PCVs. Lane M, DL2000 DNA marker; lane 1, PCV1; lane 2, PCV2; lane 3, PCV3; lane 4, TTSuV1; lane 5, TTSuV2; lane 6, PRV; lane 7, PPV; lane 8, RV; lane 9, JEV; lane 10,PEDV; lane 11, PDCoV; lane 12, ddH2O
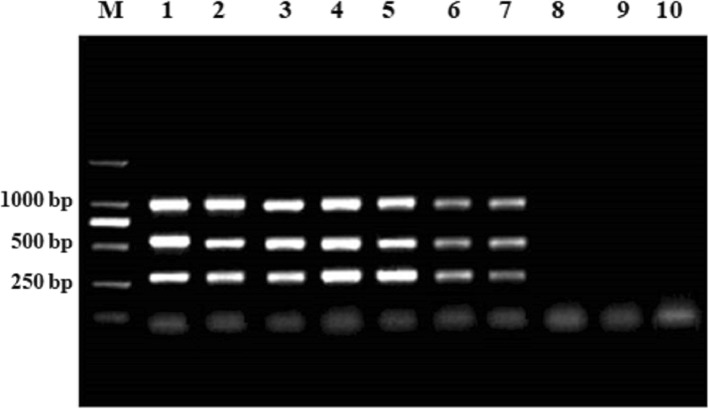
Sensitivity of the proposed multiplex PCR
The sensitivity of the proposed multiplex PCR assay was defined as the minimum DNA molecule concentration which could be detected. DNA standards, which were10-fold diluted, with known copy numbers (107 copies/μL to 10− 2 copies/μL) were used for the multiplex PCR. As shown in Fig. 4, the detection limit of the multiplex PCR was 10 copies/μL plasmid DNA molecules for PCV1, PCV2 or PCV3, which indicated that the sensitivity of the multiplex PCR was 10 copies/μL for PCV1, PCV2 and PCV3.
Fig. 4.
Sensitivity of multiplex PCR for the detection of PCVs. Lane M, DL2000 DNA marker; lanes 1–10 are: 1, 107; 2, 106; 3, 105; 4, 104; 5, 103; 6, 102; 7, 101; 8, 100; 9, 10− 1; 10, 10− 2 copies/μL
Interference test of the multiplex PCR
A mixture of different concentrations of the three PCV strains was amplified using the multiplex PCR. The results showed that detection of the three PCV strains was not influenced if the concentration of them was similar (Table 2).
Table 2.
Results of interference test of the multiplex PCR
| Virus concentration (copies/μL) | Multiplex PCR results | |||||
|---|---|---|---|---|---|---|
| PCV1 | PCV2 | PCV3 | PCV1 | PCV2 | PCV3 | |
| 102 | 101 | 100 | + | + | – | |
| 101 | 100 | 10−1 | + | – | – | |
| 100 | 10−1 | 10−2 | – | – | – | |
| 10− 1 | 10− 2 | 102 | – | – | + | |
| 102 | 102 | 102 | + | + | + | |
| 101 | 101 | 101 | + | + | + | |
| 100 | 100 | 100 | – | – | – | |
| 10−1 | 10−1 | 10−1 | – | – | – | |
+, positive result for multiplex PCR test; −, negative result for multiplex PCR test
Detection of viruses in clinical specimens
A total 286 clinical samples were tested by the multiplex PCR assay. The results were as follows: The PCV1-positive, PCV2-positive and PCV3-positive rate at the farm level was 62.5% (5/8), 87.5% (7/8) and 62.5% (5/8), respectively. The positive rates of PCV1, PCV2 and PCV3 in these samples were 52.4% (150/286), 61.2% (175/286) and 45.1% (129/286), respectively, which were 100% consistent with the sequencing method and the real-time PCR methods (Table 3). The results of the multiplex PCR method and subsequent sequencing further demonstrated the accuracy of the developed assay.
Table 3.
Detection of clinical specimens by multiplex PCR, sequencing method and real-time PCR
| Pig Farm | No. of specimens | multiplex PCR | sequencing method | real-time PCR | Concordance rate (%) |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| PCV1 positive (%) |
PCV2 positive (%) |
PCV3 positive (%) |
PCV1 positive (%) |
PCV2 positive (%) |
PCV3 positive (%) |
PCV1 positive (%) |
PCV2 positive (%) |
PCV3 positive (%) |
|||
| 1 | 50 | 0 (0) | 36 (72.0) | 33 (66.0) | 0 (0) | 36 (72.0) | 33 (66.0) | 0 (0) | 36 (72.0) | 33 (66.0) | 100 |
| 2 | 48 | 41 (85.4) | 33 (68.8) | 31 (64.6) | 41 (85.4) | 33 (68.8) | 31 (64.6) | 41 (85.4) | 33 (68.8) | 31 (64.6) | 100 |
| 3 | 38 | 35 (92.1) | 25 (65.8) | 0 (0) | 35 (92.1) | 25 (65.8) | 0 (0) | 35 (92.1) | 25 (65.8) | 0 (0) | 100 |
| 4 | 36 | 32 (88.89) | 27 (75.0) | 26 (72.2) | 32 (88.89) | 27 (75.0) | 26 (72.2) | 32 (88.89) | 27 (75.0) | 26 (72.2) | 100 |
| 5 | 33 | 0 (0) | 21 (63.6) | 20 (60.6) | 0 (0) | 21 (63.6) | 20 (60.6) | 0 (0) | 21 (63.6) | 20 (60.6) | 100 |
| 6 | 30 | 0 (0) | 17 (56.7) | 19 (63.3) | 0 (0) | 17 (56.7) | 19 (63.3) | 0 (0) | 17 (56.7) | 19 (63.3) | 100 |
| 7 | 26 | 20 (76.9) | 0 (0) | 0 (0) | 20 (76.9) | 0 (0) | 0 (0) | 20 (76.9) | 0 (0) | 0 (0) | 100 |
| 8 | 25 | 22 (88.0) | 16 (64.0) | 0 (0) | 22 (88.0) | 16 (64.0) | 0 (0) | 22 (88.0) | 16 (64.0) | 0 (0) | 100 |
| Total | 286 | 150 (52.4) | 175 (61.2) | 129 (45.1) | 150 (52.4) | 175 (61.2) | 129 (45.1) | 150 (52.4) | 175 (61.2) | 129 (45.1) | 100 |
Additionally, the PCV1 and PCV2 co-infection rate was 11.2% (32/286), the PCV1 and PCV3 co-infection rate was 5.9% (17/286), the PCV2 and PCV3 co-infection rate was 23.4% (67/286), and the PCV1, PCV2 and PCV3 co-infection rate was 1.7% (5/286), respectively in the samples from eight pig farms (Table 4). The total co-infection rate was 42.3% (121/286) in all of the detected samples.
Table 4.
Detection of the co-infection of clinical specimens by multiplex PCR
| Pig Farm | No. of specimens | PCV1 and PCV2 positive (%) |
PCV1 and PCV3 positive (%) |
PCV2 and PCV3 positive (%) |
PCV1, PCV2 and PCV3positive (%) |
|
|---|---|---|---|---|---|---|
| 1 | 50 | 0 (0) | 0 (0) | 20 (40.0) | 0 (0) | |
| 2 | 48 | 13 (27.1) | 9 (18.8) | 19 (39.6) | 2 (4.2) | |
| 3 | 38 | 9 (23.7) | 0 (0) | 0 (0) | 0 (0) | |
| 4 | 36 | 8 (22.2) | 5 (13.9) | 13 (36.1) | 3 (8.3) | |
| 5 | 33 | 0 (0) | 0 (0) | 5 (15.2) | 0 (0) | |
| 6 | 30 | 0 (0) | 0 (0) | 10 (33.3) | 0 (0) | |
| 7 | 26 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | |
| 8 | 25 | 2 (8.0) | 3 (12.0) | 0 (0) | 0 (0) | |
| Total | 286 | 32 (11.2) | 17 (5.9) | 67 (23.4) | 5 (1.7) |
Detection of viruses in animals infected with PCV
The samples from the 20 pigs were tested by the multiplex PCR assay. The results were as follows: The PCV1-positive rate was 100% (5/5) in PCV1 group, the PCV2-positive rate was 100% (5/5) in PCV2 group, and the PCV3-positive rate was 100% (5/5) in PCV3 group, respectively. None has clinical signs in PCV1 group, while two fifths in PCV2 group and there fifths in PCV3 group present with clinical symptoms as dermatitis, respiratory symptoms, severe weight loss etc. (Table 5). The results showed that the developed assay can test the viruses in the samples from “non-clinical” pigs, and indicated that it is adaptable for testing field samples.
Table 5.
Detection of the specimens from the infection pigs by multiplex PCR
| Group | No. of specimens | PCV1 positive (%) |
PCV2 positive (%) |
PCV3 positive (%) |
Clinical Signs | |
|---|---|---|---|---|---|---|
| PCV1 | 5 | 100 | 0 | 0 | none | |
| PCV2 | 5 | 0 | 100 | 0 | dermatitis, respiratory symptoms, severe weight loss (2/5) | |
| PCV3 | 5 | 0 | 0 | 100 | dermatitis, respiratory symptoms, severe weight loss (3/5) | |
| Control | 5 | 0 | 0 | 0 | none |
Discussion
PCV3 is associated with nephropathy syndrome, reproductive failure and porcine dermatitis [8], respiratory disease complex, and cardiac and multisystemic inflammation [9]. PCV2 is clinically characterized by decreased weight gain, wasting, dyspnea, and enlarged lymph nodes. It has also been identified from diseased pigs with various other clinical presentations, such as porcine dermatitis and nephropathy syndrome, porcine respiratory disease complex (PRDC) and reproductive failure [24]. PCV2 is confirmed as an endemic viral disease of swine all over the world. And in modern swine production, it is also recognized as one of the most economically important infectious pathogens [25]. Although PCV1-seroprevalence at herd level diversifies between 10 and 100% [26], it is generally believed that PCV1 has no pathogenicity to pigs [4, 6]. It was found originally as a contaminant of the porcine kidney cell line PK15 [3]. However, PCV1 has recently gained focus of attention because it was discovered as contaminant in several live veterinary vaccines and human vaccines [18, 27]. And PCV1 has been identified in cases of congenital tremors in aborted/stillborn piglets and newborn pigs. The results showed that PCV1 can proliferate and may produce pathological change in the lungs of fetal porcine [5]. And a new type of porcine circovirus in swine, a type 1 and type 2 PCV recombinant was isolated from swine samples [28]. There could be a potential damage to the piglets’ immune system caused by PCV1 infection [29]. Both PCV1 and PCV2 infections are common in pig herds worldwide [30], and PCV3 is already widely outbroken and distributed on pig farms in many countries [2, 8, 12, 13]. Single infection of PCV2 or PCV3, or co-infection of PCV1, PCV2 and PCV3 could cause many kinds of diseases in pig herds [2, 10, 13]. Veterinary workers should develop suitable prevention and control policies for PCVs and their novel strains emerging from viral evolution [31].
In order to establish prevention and control strategies for PCVs, a convenient and sensitive diagnostic method is necessary to simultaneously detect and discriminate PCVs in clinical samples. PCR assays are sensitive methods to detect a circovirus infection in viremia animals [32]. And The multiplex PCR assay is specific, rapid and easy to interpret the prevalence, epidemiology and infectious potential of some pathogens [33]. Several different PCR methods, including digital droplet PCR (ddPCR), real-time PCR and quantitative PCR (qPCR) using specific primers have been developed for detection of PCV1 and PCV2 [34–43]. And recently, two qPCR methods were proposed to detect and quantify PCV3 DNA [8, 44]. However, there is no specific, sensitive and reliable single assay capable of detecting and differentiating infection by PCV1, PCV2 and PCV3. Therefore, a specificand sensitive multiplex PCR assay to detect and discriminate PCV1, PCV2 and PCV3 in clinical specimens was developed in the present study.
In this study, an efficient and sensitive multiplex PCR assay was developed to detect and discriminate PCV1, PCV2 and PCV3 using three specific primer pairs. The sizes of the amplified PCR products of PCV1, PCV2 and PCV3 strain from the specific primers are very different, which can be easily differentiated by electrophoresis. The specific primers for the multiplex PCR assay successfully amplified the 310-bp PCV1, 505-bp PCV2 or 1021-bp PCV3 gene. Furthermore, the multiplex PCR assay with three sets of PCV1-, PCV2- and PCV3-specific primers simultaneously detected and discriminated PCV1, PCV2 and PCV3 DNA in a single reaction. And the sensitivity of the multiplex PCR assay was 10 copies/μL for PCV1, PCV2 and PCV3, which is similar to several previous studies [2, 34, 44, 45].
Although multiplex qPCR assays that can discriminate PCV2 and PCV3 have already been developed, including one study from China [46], but all the assays must be run in an expensive instrument—Fluorescent Quantatitive PCR. It is unaffordable for many laboratories especially those in the counties and towns in China, so the multiplex qPCR assays couldn’t be run in their laboratories. However, almost all of them are equipped with the conventional PCR instrumentation. The sensitivity of the multiplex PCR developed in this study is similar to several qPCR assays. Definitely, the multiplex PCR is a convenient and sensitive diagnostic method to detect and discriminate PCVs in clinical samples in China and any other developing countries.
A total of 286 specimens from eight pig farms in Hubei province, central China, were analyzed using the proposed multiplex PCR. The results were 100% same as those of sequencing method and real-time PCR methods. The PCV1, PCV2 and PCV3 singular infection rate, the PCV1 and PCV2 or PCV3 co-infection rate, the PCV2 and PCV3 or PCV1, PCV2 and PCV3 co-infection rate were higher than the previous reports [2, 16, 45]. The results indicated that the PCVs infection and their co-infection are severe in Hubei Province, Central China. And the epidemiology and genome characterizations of PCVs in Hubei Province would be further studied in the near future.
DNAs of PCV2 and PCV3 were detected using multiplex PCR in aborted fetal tissue samples and respiratory diseased piglet tissue samples. The results suggested that both PCV2 and PCV3 infection are associated with reproductive failure and respiratory disease at the infection pig farms, as previous researches [2, 8, 44]. The singular infection of PCV2 and PCV3, and PCV2, PCV3 and/or PCV1 co-infection play an etiological role in porcine circovirus associated disease (PCVAD), which had caused huge economic losses to pig farms all over the world.
The multiplex PCR assay is sensitive, and it can also simultaneously discriminate PCV1, PCV2, and PCV3 in a single reaction, which makes it lower cost and less time. It will be a useful tool to detect and discriminate PCVs in field samples. Considering the prevalence of PCV1, PCV2 and PCV3 co-infection in the field, the multiplex PCR will enable the correct diagnosis of suspected clinical cases and stimulate further epidemiological researches for its control. However, there would be any other PCV genotypes in pig farms. Hence, an accurater PCV identification which covers all types of PCV genotypes should be established in the near future.
Conclusions
In conclusion, the developed multiplex PCR is a convenient, sensitive, efficient, and highly specific assay to detect and discriminate PCVs, which will be useful in etiological and epidemiological studies, as well as diagnosis in clinical cases. The accuracy and simplicity of the assay makes it a useful, suitable and powerful tool for PCVs detection, prevention and control in China and any other developing countries.
Acknowledgements
We would like to thank Professor Kemei Peng, College of Animal Science and Veterinary Medicine, Huazhong Agricultural University for reviewing the manuscript.
Abbreviations
- JEV
Japanese encephalitis virus
- PCV
Porcine circovirus
- PCV1
Porcine circovirus type 1
- PCV2
Porcine circovirus type 2
- PCV3
Porcine circovirus type 3
- PDCoV
Porcine deltacorona virus
- PEDV
Porcine epidemic diarrhea virus
- PPV
Porcine parvovirus
- PRV
Pseudorabies virus
- RV
Rotavirus
- TTSuV1
Torque teno sus viruse type 1
- TTSuV2
Torque teno sus viruse type 2
Author contributions
KLY and YXT conceived and designed the experiments. ZWJ, RG, KLY and ZYD performed the experiments. KLY, ZWJ and DNZ analyzed the data. KLY wrote the paper. All authors read and approved the final manuscript.
Funding
The study was supported by the National Key R & D Program of China (2016YFD0500703), the Opening Subject of Key Laboratory of Prevention and Control Agents for Animal Bacteriosis (Ministry of Agriculture) (2017ZD06). It was also financed by Hubei Province Innovation Center of Agricultural Sciences and Technology (2016–620–000-001-026). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
This study was approved by the Animal Ethics Committee of the Hubei province and all subjects provided written informed consent.
Consent for publication
Not applicable.
Competing interests
The authors have declared that no competing interests exist.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Keli Yang, Email: keliy6@126.com.
Zuwu Jiao, Email: hbxms@126.com.
Danna Zhou, Email: zdn_66@126.com.
Rui Guo, Email: hlguorui@163.com.
Zhengying Duan, Email: zy001d@sina.com.
Yongxiang Tian, Email: tyxanbit@163.com.
References
- 1.Mankertz A, Domingo M, Folch JM, LeCann P, Jestin A, Segalés J, Chmielewicz B, Plana-Durán J, Soike D. Characterisation of PCV-2 isolates from Spain, Germany and France. Virus Res. 2000;66:65–77. doi: 10.1016/S0168-1702(99)00122-7. [DOI] [PubMed] [Google Scholar]
- 2.Ku X, Chen F, Li P, Wang Y, Yu X, Fan S, Qian P, Wu M, He Q. Identification and genetic characterization of porcine circovirus type 3 in China. Transbound Emerg Dis. 2017;64:703–708. doi: 10.1111/tbed.12638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tischer I, Rasch R. Tochtermann, G. Characterization of papovavirus-and picornavirus-like particles in permanent pig kidney cell lines. Zentralbl Bakteriol Parasitenkd Infekionskr Hyg, I Abt Orig,reihe A Med Mikrobiol Parasitol, 226(1974) 153–167. [PubMed]
- 4.Tischer I, Mields W, Wolff D, Vagt M, Griem W. Studies on epidemiology andpathogenicity of porcine circovirus. Arch Virol. 1986;91(3–4):271–276. doi: 10.1007/BF01314286. [DOI] [PubMed] [Google Scholar]
- 5.Saha D, Lefebvre DJ, Ducatelle R, Doorsselaere JV, Nauwynck HJ. Outcome of experimental porcine circovirus type 1 infections in mid-gestational porcine foetuses. BMC Vet Res. 2011;7:64. doi: 10.1186/1746-6148-7-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Allan GM, McNeilly F, Kennedy S, Daft B, Clarke EG, Ellis JA, Haines DM, Meehan BM, Adair BM. Isolation of porcine circovirus-like viruses from pigs with a wasting disease in the USA and Europe. J Vet Diagn Invest. 1998;10:3–10. doi: 10.1177/104063879801000102. [DOI] [PubMed] [Google Scholar]
- 7.Opriessnig T, Meng XJ, Halbur PG. Porcine circovirus type 2 associated disease: update on current terminology, clinical manifestations, pathogenesis, diagnosis, and intervention strategies. J Vet Diagn Invest. 2007;19:591–615. doi: 10.1177/104063870701900601. [DOI] [PubMed] [Google Scholar]
- 8.Palinski R, Piñeyro P, Shang P, Yuan F, Guo R, Fang Y, Byers E, Hause BM. A novelporcine circovirus distantly related to known circoviruses is associated with porcine dermatitis and nephropathy syndrome and reproductive failure. J Virol. 2017;91:e01879–e01816. doi: 10.1128/JVI.01879-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Phan TG, Giannitti F, Rossow S, Marthaler D, Knutson T, Li L, Deng X, Resende T, Vannucci F, Delwart E. Detection of a novel circovirus PCV3 in pigs with cardiac and multi-systemic inflammation. Virol J. 2016;13(1):184. doi: 10.1186/s12985-016-0642-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collins PJ, McKillen J, Allan G. Porcine circovirus type 3 inthe UK. Vet Rec. 2017;181(22):599. doi: 10.1136/vr.j5505. [DOI] [PubMed] [Google Scholar]
- 11.Stadejek T, Wozniak A, Miłek D, Biernacka K. First detection of porcine circovirus type 3 on commercial pig farms in Poland. Transbound Emerg Dis. 2017;64:1350–1353. doi: 10.1111/tbed.12672. [DOI] [PubMed] [Google Scholar]
- 12.Franzo G, Legnardi M, Hjulsager CH, Klaumann F, Larsen LE, Segales J, Drigo M. Full genome sequencing of porcine circovirus 3 fields strains from Denmark, Italy and Spain demonstrates and high within-Europe genetic heterogenecity. Transbound Emerg Dis. 2018;65(3):602–606. doi: 10.1111/tbed.12836. [DOI] [PubMed] [Google Scholar]
- 13.Kwon T, Yoo SJ, Park CK, Lyoo YS. Prevalence ofnovel porcine circovirus 3 in Korean pig populations. Vet Microbiol. 2017;207:178–180. doi: 10.1016/j.vetmic.2017.06.013. [DOI] [PubMed] [Google Scholar]
- 14.Tochetto C, Lima DA, Varela APM, Loiko MR, Paim WP, Scheffer CM, Herpich JI, Cerva C, Schmitd C, Cibulski SP, Santos AC, Mayer FQ, Roehe PM. Full-Genome Sequence of Porcine Circovirus type 3 recovered from serum of sows with still birthsin Brazil. Transbound Emerg Dis. 2018;65(1):5–9. doi: 10.1111/tbed.12735. [DOI] [PubMed] [Google Scholar]
- 15.Fan S, Ku X, Chen F, Wang Y, Yu X, He Q. Complete genome sequence of a novel porcine circovirus type 3 strain, PCV3/CN/Hubei-618/2016, Isolated from China. Genome Announc. 2017;5(15):e00100–e00117. doi: 10.1128/genomeA.00100-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhai SL, Zhou X, Zhang H, Hause BM, Lin T, Liu R, Chen QL, Wei WK, Lv DH, Wen XH, Li F, Wang D. Comparative epidemiology of porcine circovirus type 3 in pigs with different clinical presentations. Virol J. 2017;14(1):222. doi: 10.1186/s12985-017-0892-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng S, Wu X, Zhang L, Xin C, Liu Y, Shi J, Peng Z, Xu S, Fu F, Yu J, Sun W, Xu S, Li J, Wang J. The occurrence of porcine circovirus 3 without clinical infection signs in Shandong Province. Transbound Emerg Dis. 2017;64(5):1337–1341. doi: 10.1111/tbed.12667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beach NM, Juhan NM, Cordoba L, Meng XJ. Replacement of the replication factors of porcine circovirus (PCV) type 2 with those of PCV type 1 greatly enhances viral replication in vitro. J Virol. 2010;84:8986–8989. doi: 10.1128/JVI.00522-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li WJ. Molecular Epidemiology of Porcine Cireovirus Type 2 Isolates Collected in Some Area of China (in Chinese). D. Thesis (2009) Huazhong Agricultural University. Wuhan, China.
- 20.Yang KL, Tian YX, Zhou DN, Duan ZY, Guo R, Liu ZW, Yuan FY, Liu W. A multiplex RT-PCR assay to detect and discriminate porcine reproductive and respiratory syndrome viruses in clinical specimens. Viruses. 2017;9:205. doi: 10.3390/v9080205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parida M, Shukla J, Sharma S, Santhosh SR, Ravi V, Mani R, Thomas M, Khare S, Rai A, Ratho RK, Pujari S, Mishra B, Rao PVL, Vijayaraghavan R. Development and evaluation of reverse transcription loop-mediated isothermal amplification assay for rapid and real-time detection of the swine-origin influenza a H1N1 virus. J Mol Diagn. 2011;13:100–107. doi: 10.1016/j.jmoldx.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim HR, Park YR, Lim D, Park MJ, Park JY, Kim SH, Lee KK, Lyoo YS, Park CK. Multiplex real-time polymerase chain reaction for the differential detection of porcine circovirus 2 and 3. J Virol Methods. 2017;250:11–16. doi: 10.1016/j.jviromet.2017.09.021. [DOI] [PubMed] [Google Scholar]
- 23.Li J., Shi J-l., Wu X-y., Cong X-y., Xu S-j., Yuan X-y., Wu J-q., Sun W-b., Du Y-j., Peng Z., Wang J-b., Huang B-h. Differentiation of PCV1 and PCV2 by a multiplex real-time PCR assay. Veterinary Record. 2013;173(14):346.1-346. doi: 10.1136/vr.101686. [DOI] [PubMed] [Google Scholar]
- 24.Chen DJ, Wei YW, Huang LP, Wang YP, Sun JH, Du WJ, Wu HL, Liu CM. Synergistic pathogenicity in sequential coinfection with Mycoplasma hyorhinis and porcine circovirus type 2. Vet Microbiol. 2016;182:123–130. doi: 10.1016/j.vetmic.2015.11.003. [DOI] [PubMed] [Google Scholar]
- 25.Opriessnig T, Madson DM, Prickett JR, Kuhar D, Lunney JK, ElsenerJ HPG. Effect of porcine circovirus type 2 (PCV2) vaccination on porcine reproductive and respiratory syndrome virus (PRRSV) and PCV2 coinfection. Vet Microbiol. 2008;131(1–2):103–114. doi: 10.1016/j.vetmic.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 26.Puvanendiran S, Stone S, Yu W, Johnson CR, Abrahante J, Jimenez LG, Griggs T, Haley C, Wagner B, Murtaugh MP. Absence of porcine circovirus type 1 (PCV1) and high prevalence of PCV 2 exposure and infection in swine finisher herds. Virus Res. 2011;157(1):92–98. doi: 10.1016/j.virusres.2011.02.012. [DOI] [PubMed] [Google Scholar]
- 27.Pastoret PP. Human and animal vaccine contaminations. Biologicals. 2010;38:332–334. doi: 10.1016/j.biologicals.2010.02.015. [DOI] [PubMed] [Google Scholar]
- 28.Gagnon CA, Music N, Fontaine G, Tremblay D, Harel J. Emergence of a new type of porcine circovirus in swine (PCV): a type 1 and type 2 PCV recombinant. Vet Microbiol. 2010;144(1–2):18–23. doi: 10.1016/j.vetmic.2009.09.072. [DOI] [PubMed] [Google Scholar]
- 29.Cao L, Sun W, Lu H, Tian M, Xie C, Zhao G, Han J, Wang W, Zheng M, Du R, Jin N, Qian A. Genetic variation analysis of PCV1 strains isolated from Guangxi Province of China in 2015. BMC Vet Res. 2018;14(1):43. doi: 10.1186/s12917-018-1345-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Allan G, Krakowka S, Ellis J, Charreyre C. Discovery and evolving history oftwo genetically related but phenotypically different viruses, porcine circoviruses 1 and 2. Virus Res. 2012;64:4–9. doi: 10.1016/j.virusres.2011.09.013. [DOI] [PubMed] [Google Scholar]
- 31.Liu JK, Wei CH, Ailing Dai AL, Lin ZF, Fan KW, Fan JL, Liu JY, Luo ML, Yang XY. Detection of PCV2e strains in Southeast China. Peer J. 2018;6:e4476. doi: 10.7717/peerj.4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Denner J, Mankertz A. Porcine Circoviruses and Xenotransplantation. 2017;Viruses, 9:83. 10.3390/v9040083. [DOI] [PMC free article] [PubMed]
- 33.Harmon KM, Wesley IV. Multiplex PCR for the identification of Arcobacter and differentiation of Arcobacter butzleri from other arcobacters. Vet Microbiol. 1997;58(2–4):215–227. doi: 10.1016/S0378-1135(97)00151-X. [DOI] [PubMed] [Google Scholar]
- 34.Brunborg IM, Moldal T, Jonassen CM. Quantitation of porcine circovirus type2 isolated from serum/plasma and tissue samples of healthy pigs and pigs with postweaning multisystemic wasting syndrome using a TaqMan-based real-time PCR. J Virol Methods. 2004;122:171–178. doi: 10.1016/j.jviromet.2004.08.014. [DOI] [PubMed] [Google Scholar]
- 35.Chang GN, Huang JF, Chen JT, Tsen HY, Wang JJ. Fast diagnosis and quantification for porcine circovirus type 2 (PCV-2) using real-time polymerase chain reaction. J Microbiol Immunol Infect. 2010;43:85–92. doi: 10.1016/S1684-1182(10)60014-X. [DOI] [PubMed] [Google Scholar]
- 36.Olvera A, Sibila M, Calsamiglia M, Segalés J, Domingo M. Comparison of porcine circovirus type 2 load in serum quantified by a real time PCR in post weaning multisystemic wasting syndrome and porcine dermatitis and nephropathy syndrome naturally affected pigs. J Virol Methods. 2004;117:75–80. doi: 10.1016/j.jviromet.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 37.Quintana J, Segalés J, Calsamiglia M, Domingo M. Detection of porcine circovirus type 1 in commercial pig vaccines using polymerase chain reaction. Vet J. 2006;171:570–573. doi: 10.1016/j.tvjl.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 38.Rovira A, Balasch M, Segales J, Garcia L, Plana-Duran J, Rosell C, Ellerbrok H, Mankertz A, Domingo M. Experimental inoculation of conventional pigswith porcine reproductive and respiratory syndrome virus and porcine circovirus 2. J Virol. 2002;76:3232–3239. doi: 10.1128/JVI.76.7.3232-3239.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Segalés J, Calsamiglia M, Olvera A, Sibila M, Badiella L, Domingo M. Quantification of porcine circovirus type 2 (PCV2) DNA in serum and tonsillar, nasal, tracheo-bronchial, urinary and faecal swabs of pigs with and without postweaning multisystemic wasting syndrome (PMWS) Vet Microbiol. 2005;111:223–229. doi: 10.1016/j.vetmic.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 40.Wang C, Pang V, Lee F, Liao PC, Huang YL, Lin YL, Lai SS, Jeng CR. Development and evaluation of a loop-mediated isothermal amplification method for rapid detection and differentiation of two genotypes of porcine circovirus type 2. J Microbiol Immunol. 2014;47:363–370. doi: 10.1016/j.jmii.2013.05.003. [DOI] [PubMed] [Google Scholar]
- 41.Wang J, Wang J, Liu L, Li R, Yuan W. Rapid detection of Porcine circovirus 2 by recombinase polymerase amplification. J Vet Diagn Invest. 2016;28:574–578. doi: 10.1177/1040638716654201. [DOI] [PubMed] [Google Scholar]
- 42.Zhao K, Han F, Zou Y, Zhu L, Li C, Xu Y, Zhang C, Tan F, Wang J, Tao S, He X, Zhou Z, Tang X. Rapid detection of porcine circovirus type 2 using a TaqMan-based real-time PCR. Virol J. 2010;7:374. doi: 10.1186/1743-422X-7-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao S, Lin H, Chen S, Yang M, Yan Q, Wen C, Hao Z, Yan Y, Sun Y, Hu J, Chen Z, Xi L. Sensitive detection of porcine circovirus-2 by droplet digital polymerase chain reaction. J Vet Diagn Investig. 2015;27(6):784–788. doi: 10.1177/1040638715608358. [DOI] [PubMed] [Google Scholar]
- 44.Wang J, Zhang Y, Zhang R, Han Q, Wang J, Liu L, Li R, Yuan W. Recombinase polymerase amplification assay for rapid detection of porcine circovirus 3. Mol Cell Probes. 2017;36:58–61. doi: 10.1016/j.mcp.2017.09.001. [DOI] [PubMed] [Google Scholar]
- 45.Zhang L, Luo Y, Liang L, Li J, Cui S. Phylogenetic analysis of porcine circovirus type 3 and porcine circovirus type 2 in China detected by duplex nanoparticle-assisted PCR. Infect Genet Evol. 2018;60:1–6. doi: 10.1016/j.meegid.2018.02.006. [DOI] [PubMed] [Google Scholar]
- 46.Li X, Qiao M, Sun M, Tian K. A duplex real-time PCR assay for the simultaneous detection of porcine circovirus 2 and circovirus 3. Virol Sin. 2018;33(2):181–186. doi: 10.1007/s12250-018-0025-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.