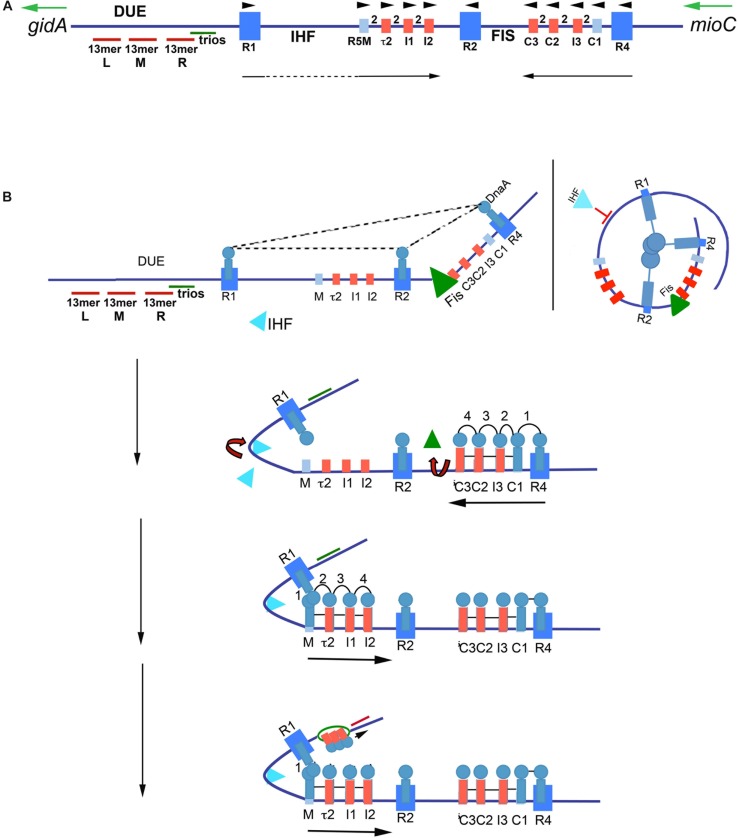
FIGURE 2.
Model of staged orisome assembly. (A) Map of E. coli oriC. High affinity R boxes are marked by large blue rectangles, low affinity DnaA-ATP recognition sites are marked by small red rectangles, and low affinity non-discriminatory recognition sites are marked by small light blue rectangles. Left (l), middle (M), and right (R) 13 mer AT-rich repeats are shown, as well as the locations of the DnaA-trio elements, and Fis and IHF binding regions. Green arrows mark direction of transcription of flanking genes gidA and mioC, black arrows mark the direction of DnaA binding progression, and black arrowheads mark the orientation of the recognition sites based on the direction faced by the arginine finger (R285) of bound DnaA. (B) Stages of orisome assembly. Stage 1 (left): After initiation of chromosome replication, DnaA rebinds to high affinity R1, R2, and R4 sites. Fis is also bound at this stage, but IHF is not. Low affinity sites are unoccupied. Dashed lines indicate interaction between bound DnaA molecules (right): Looping of DNA to allow bound DnaA molecules to interact. Stage 2: DnaA bound to R4 recruits DnaA for binding to C1. DnaA then progressively fills the remaining arrayed sites, forming an oligomer in the gap region between R2 and R4. The DnaA oligomer displaces Fis, and loss of Fis allows IHF to bind to its cognate site. Stage 3: The bend induced by IHF binding allows DnaA, recruited by R1, to bind to R5M, and form a cross-strand DnaA interaction. A DnaA oligomer then progressively grows toward R2, bound to arrayed low affinity sites, and anchored by R2. Stage 4: oriC DNA is unwound in the DUE, and DnaA in the form of a compact filament binds to the ssDNA in DnaA-trios. Figure adapted from Leonard and Grimwade (2015).