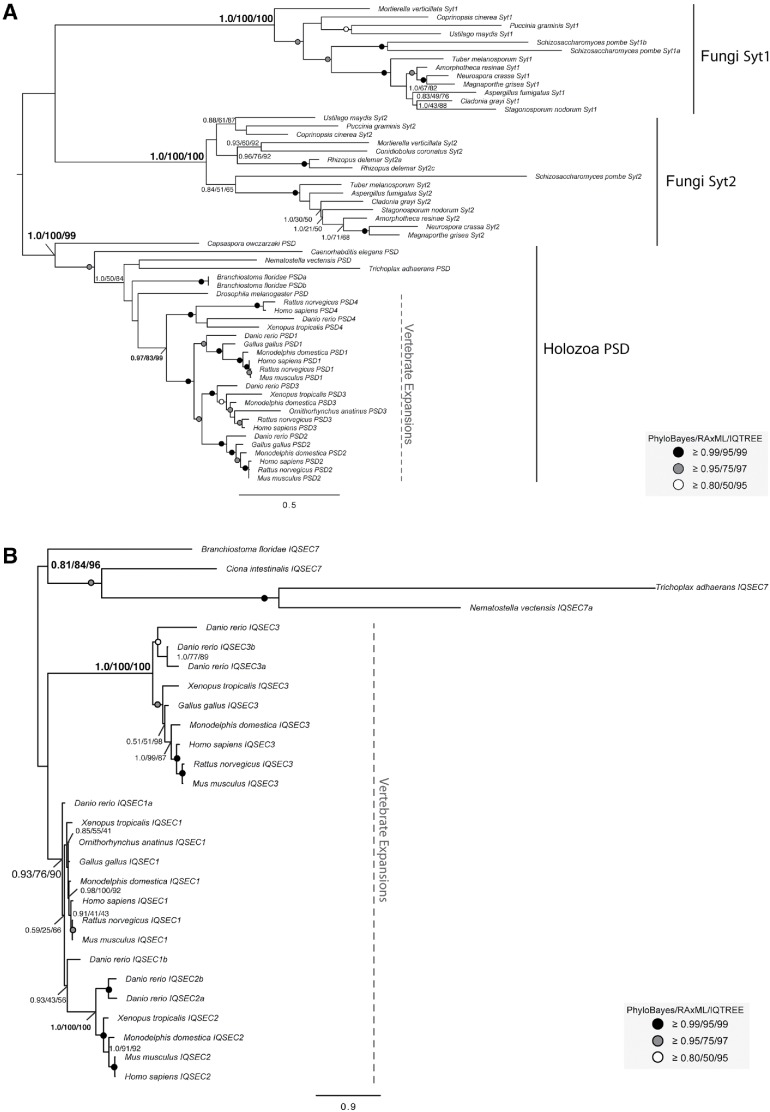
FIGURE 7:
Phylogenetic examination of holozoan cytohesin-derived family paralogues reveals multiple expansion events. (A) Phylogeny of the holozoan and fungal EFA6 proteins identify two separate clades. A duplication event in the EFA6 proteins at the base of opisthokonta yields holozoan PSD proteins and fungal Syt proteins. Further duplications in the EFA6/PSD subfamily have occurred with the transition into metazoa to yield PSD1-4 and in the fungal Syt to yield Syt1 and Syt2. (B) Phylogenetic analysis of IQSEC7 paralogues from holozoa was carried out to determine when the duplication events producing the three vertebrate paralogues occurred. The analysis reveals two duplications at the base of vertebrates, resulting in three paralogues, with IQSEC7 3/BRAG3 diverging first, followed by IQSEC7 1/BRAG1 and IQSEC7 2/BRAG2. The best Bayesian topology is shown. In both instances, numerical values represent Bayesian probabilities (PhyloBayes)/maximum-likelihood non-parametric bootstrap values (RAxML)/ultrafast bootstrap values (IQTREE). Nodes of interest are in bold. Values for other supported nodes have been replaced by symbols: closed dark circle ≥0.99/95/99, closed light circles ≥0.95/75/97, and open circles ≥0.80/50/95. The tree was rooted at filasterean IQSEC7 sequences.