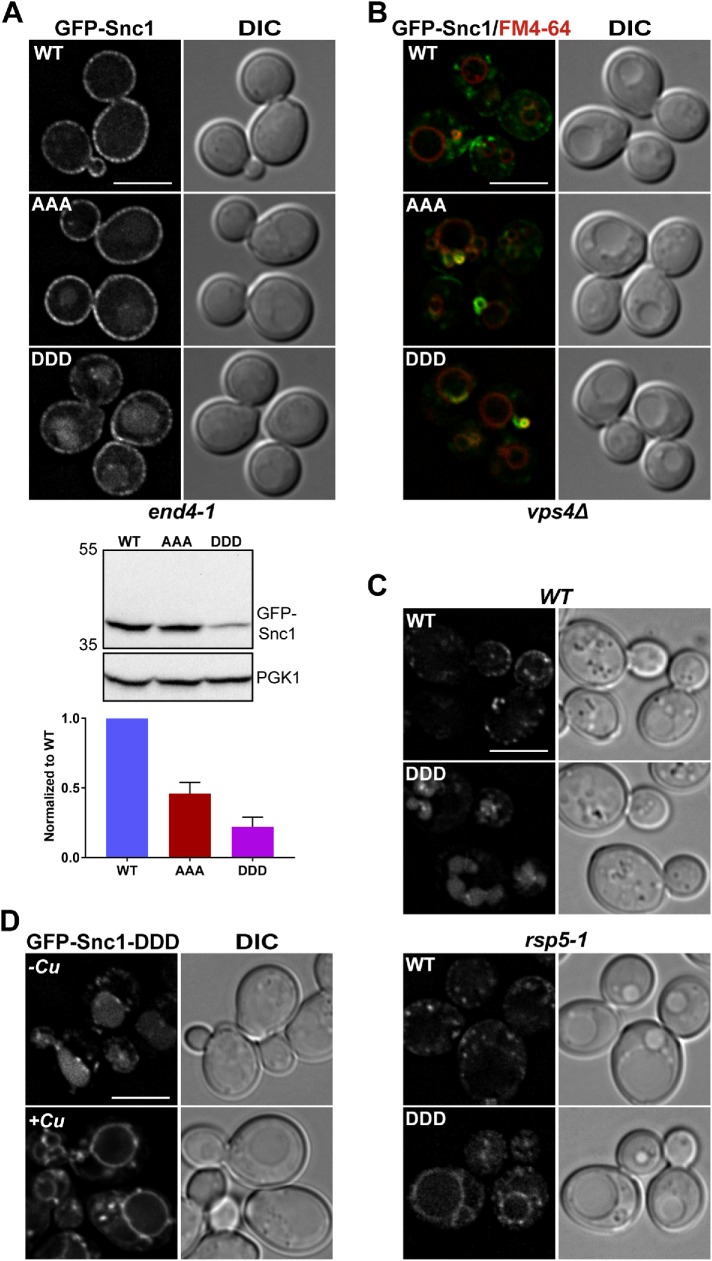
FIGURE 5:
Snc1 transmembrane domain mutants are defective for trafficking via the Snx4 retrograde pathway. (A) Micrographs of end4-1 cells expressing the indicated GFP-Snc1 constructs. Note that all GFP-Snc1 mutants tested localize to both the plasma membrane and the vacuole labeled by FM4-64. Cell lysates of end4-1 cells expressing the indicated GFP-Snc1 point mutants were probed with anti-GFP and the amount of GFP-Snc1 was determined by semiquantitative Western blot analysis. Anti-PGK1 immunoblot was used as protein loading control. The positions of molecular mass (kDa) markers are indicated. The results from three experiments were averaged and standard error of the mean indicated. (B) Micrographs of vps4Δ cells expressing indicated GFP-Snc1 constructs. Note that all GFP-Snc1 mutants tested localize to the “class E compartment” and were absent from the vacuole. Class E compartments and vacuoles were visualized using FM4-64 dye. (C) Micrographs of rsp5-1 cells and isogenic wild-type cells expressing GFP-Snc1 or GFP-Snc1-DDD. Note that GFP-Snc1-DDD localizes to the vacuole membrane instead of the vacuole lumen in rsp5-1 cells. (D) Micrographs of wild-type cells coexpressing Rsp5-DUb and GFP-Snc1-DDD. Rsp5-DUb is driven by the CUP1 promoter and to induce expression, cells were incubated with 50 μM CuCl2 for 4 h at 30°C. An approximate medial single Z section is shown unless otherwise indicated. Scale bars indicate 5 μm.