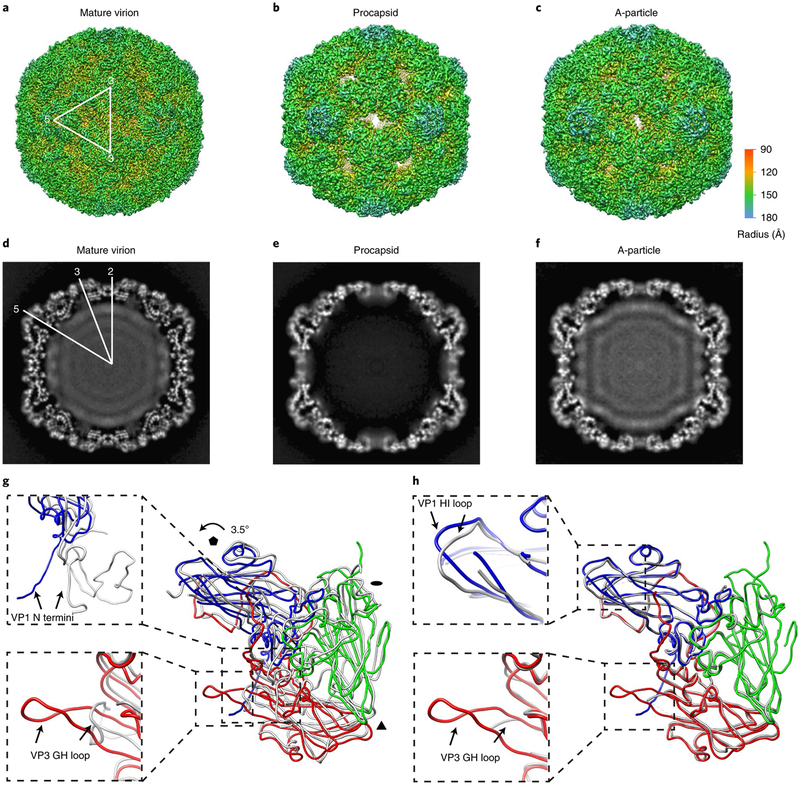
Fig. 1 |. The cryoEM structures of EV-D68 particles.
a–c, Iso-contoured views (along the icosahedral two-fold axis) of cryoEM maps (radially coloured) of the mature virion (a), procapsid (b) and A-particle (c). One icosahedral asymmetric unit is marked by a white triangle in a. The procapsid and A-particle show capsids with open two-fold channels, which are closed in the mature virion. d–f, Central sections of the corresponding maps displayed in the upper row. The densities of encapsidated genomic RNA are present in both the mature virion (d) and A-particle (f), but absent in the procapsid (e). g,h, Superpositions of protomers of A-particle protomer (VP1: blue; VP2: green; VP3: red) with that of mature virion (grey) (g) or procapsid (grey) (h). The orientation of the protomers is marked by black polygons indicating icosahedral five-, three- and two-fold axes, respectively. The colour scheme for the capsid proteins will be kept the same in all figures unless noted otherwise. The regions of the most significant differences between models are highlighted with magnified views in the dashed boxes: VP1 N termini (upper box) and VP3 GH loops (lower box) between the A-particle and the mature virion (g); VP1 HI loops (upper box) and VP3 GH loops (lower box) between the A-particle and the procapsid (h).