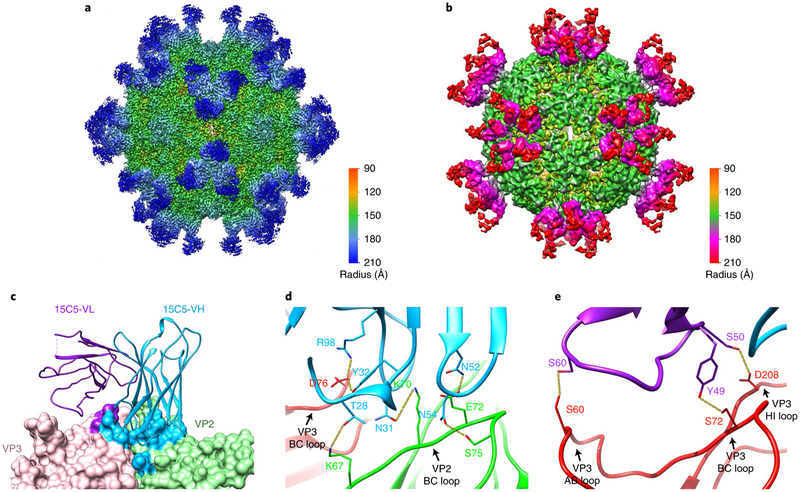
Fig. 3 |. CryoEM structures of immune complexes EV-D68-M:15C5 and EV-D68-A:11G1.
a,b, Iso-contoured views of cryoEM maps (radially coloured) of the immune complexes EV-D68-M:15C5 (a) and EV-D68-A:11G1 (b). Groups of three 15C5 Fabs and five 11G1 Fabs bind at each three-fold and five-fold axis, respectively, c, Surface representation shows the interaction interface between Fab 15C5 and the capsid. Each Fab 15C5 binds across VP2 and VP3 from two adjacent protomers. Footprints of the VH and VL of the Fab on the capsid are coloured with light blue and purple, respectively. d,e, Close-up views of the interaction interface between the capsid and either the heavy (d) or light chain (e) of Fab 15C5. Note that the heavy chain binds across the VP2 BC loop and the VP3 BC loop from two adjacent protomers as shown in d. Potential hydrogen bonds and salt bridges are marked by yellow dashed lines.