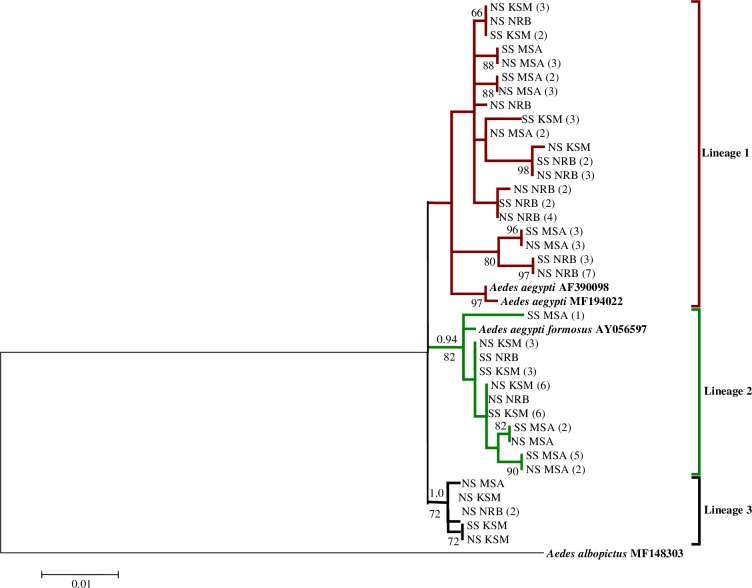
Fig 3. Maximum likelihood tree inferred using the (GTR+G) model of sequence evolution for COI barcode region (860 bp) of dengue virus susceptible (SS) and non-susceptible (NS) Aedes aegypti mosquitoes from Mombasa (MSA), Kisumu (KSM), and Nairobi (NRB), Kenya.
The number of individuals sharing a haplotype is indicated in parentheses. Bayesian posterior probabilities ≥0.90 and bootstrap support values from 5,000 replications ≥65 are indicated above and below the three major lineages, respectively, with terminal nodes reflecting bootstrap support values alone. Aedes albopictus was included for outgroup purposes.