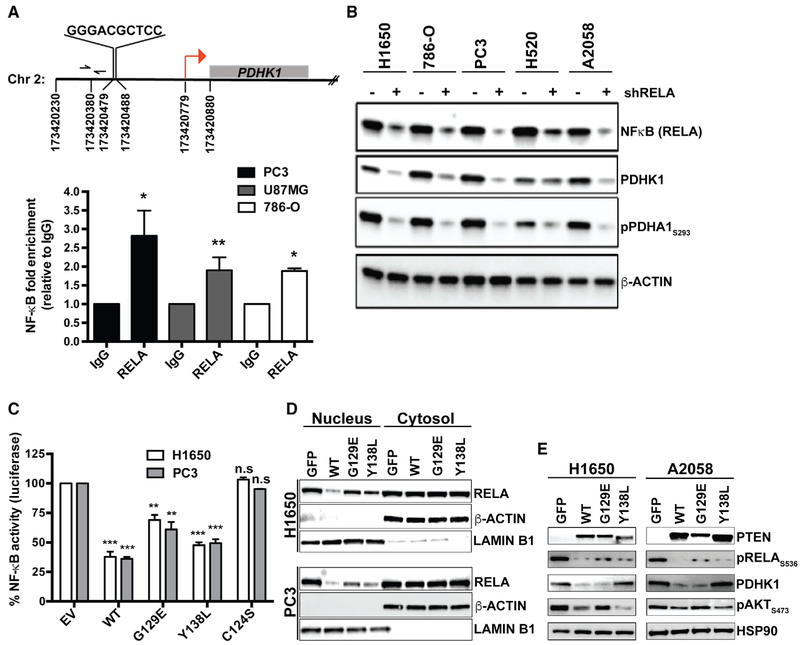
Figure 4. PTEN Protein Phosphatase Represses PDHK1 by Suppressing NF-κB Activation.
(A) Top: identification of NF-κB consensus binding site in the promoter of PDHK1 gene located between nucleotide positions 173420479 and 173420488 in chromosome 2, ~300 bp upstream of the transcription start site (TSS; red arrow) at nucleotide position 173420779. Bottom: NF-κB (RELA) recruitment at PDHK1 promoter in PTEN-deficient cancer cells by ChIP assay. Primers (forward and reverse arrows) used to amplify a 118 bp region (spanning nucleotide position 173420380) ~40 bp upstream of the NF-κB binding site in the PDHK1 promoter are shown. Fold enrichment (RELA ChIP DNA [pg] to IgG DNA [pg]) data are shown as mean ± SEM (n = 3 replicates). *p < 0.05 and **p < 0.01 compared with “IgG control” by two-tailed unpaired t test with Welch’s correction.
(B) Western blots showing NF-κB (RELA), PDHK1, and phospho-PDHA1 expression in PTEN-deficient cancer cell lines with or without stable RELA knockdown. shRELA, shRNA to RELA.
(C) Effects of PTENWT or PTENG129E or PTENY138L or empty vector (EV) expression in PTEN-deficient cancer cell lines on NF-κB activity by luciferase reporter assays (STAR Methods) are shown. Data are shown as mean ± SD (n = 2 replicates). **p < 0.01 and ***p < 0.001 compared with “empty vector control expressing PTEN-deficient cells” by Tukey’s multiple-comparisons one-way ANOVA.
(D) Effects of PTENWT or PTENG129E or PTENY138L or GFP expression in PTEN-deficient cancer cell lines on NF-κB (RELA) subcellular localization by nuclear-cytoplasmic fractionation and immunoblotting are shown. Western blots were also probed with anti-LaminB1 and anti-actin β antibodies as nuclear and cytoplasmic markers, respectively.
(E) Western blots showing PTEN, phospho-RELA, PDHK1, and phospho-AKT expression in PTEN-deficient cancer cell lines stably expressing PTENWT, PTENG129E, PTEN138L, or GFP. See also Figure S8 and Table S3.