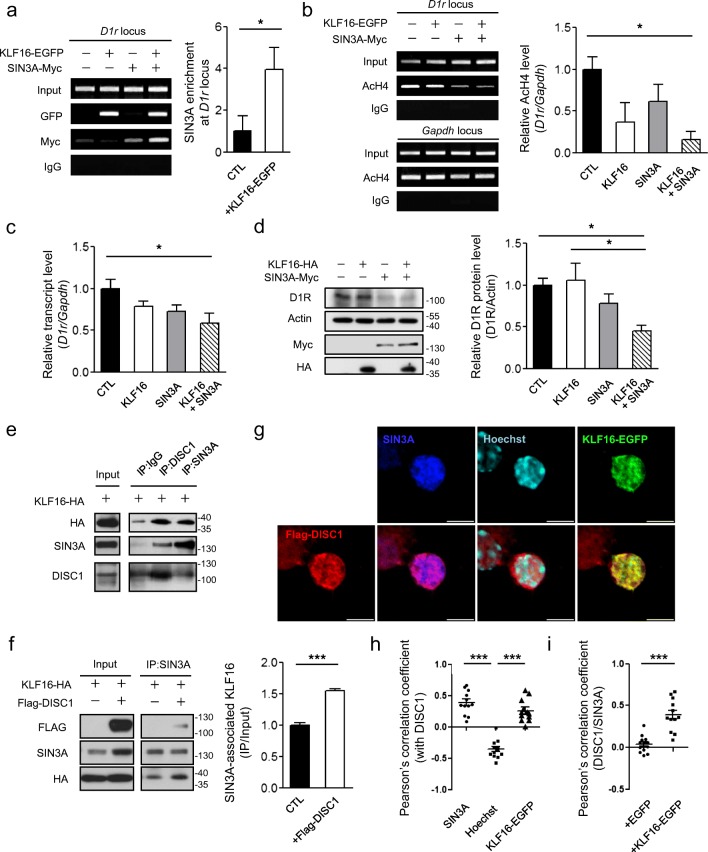
Fig. 3.
Recruitment of SIN3A corepressor by the KLF16-DISC1 complex at the D1r locus. a Enhanced ChIP-PCR signal of SIN3A-Myc at the D1r locus upon co-expression of KLF16 in differentiated CAD cells (n = 3). Antibody against GFP and Myc were used for IP. b Reduced acetylated H4 level at the D1r locus upon KLF16 and SIN3A overexpression in differentiated CAD cells (n = 3). Anti-AcH4 (K5, K8, K12, and K16) antibody was used for IP. c Reduced transcript abundance of D1r upon KLF16 and SIN3A overexpression in differentiated CAD cells (n = 6). d Reduced endogenous D1R protein level upon KLF16 and SIN3A overexpression in differentiated CAD cells (n = 4). e CoIP of KLF16-HA with endogenous DISC1 and SIN3A from differentiated CAD cell lysate. f Enhanced coIP of KLF16-HA with endogenous SIN3A upon Flag-DISC1 overexpression in differentiated CAD cells (n = 3). Anti-SIN3A antibody was used for IP. g Confocal images of Flag-DISC1 (red, left bottom), SIN3A (blue), Hoechst (cyan), KLF16-EGFP (green), and their merged images (bottom) of the nucleus of cultured mouse striatal neuron at DIV10. Scale bars represent 5 μm. h Pearson’s correlation coefficients between DISC1 and SIN3A, between DISC1 and Hoechst, and between DISC1 and KLF16-EGFP (n = 12). i Effect of KLF16-EGFP overexpression on the Pearson’s correlation coefficient between Flag-DISC1 and SIN3A (n = 14 for EGFP; n = 12 for KLF16-EGFP). Confocal images were obtained from at least three separate coverslips. Deconvoluted images and Pearson’s correlation coefficients were obtained by the CellSens software (Olympus). *P < 0.05; ***P < 0.001; one-way ANOVA with post-hoc Tukey test (b, c, d, h); two-tailed t test (a, f, i)