Abstract
Staphylococcus aureus is an opportunistic bacterium which is carried as a normal flora organism but has a major role in the epidemiology and pathogenesis of different staphylococcal infections in humans and animals. Fifty S. aureus isolated from banknotes, foods, human infections and bovine mastitis were subjected to DNA fingerprinting by randomly amplified polymorphic DNA (RAPD) analysis to examine their genotypic polymorphism and investigate the amount of genetic relatedness among these various isolates. At 100% RAPD profile similarity level, isolates were classified into four, five and seven groups of the same clone, according to the RAPDPCR with OLP6, OLP11 and OLP13 primers, respectively. Amplification of the isolates resulted in several polymorphic bands ranged from >50 to >1500 bp in size. Maximum number of bands was obtained by primer OLP13 which produced seven bands in bovine mastitis isolates. Most polymorphisms were observed in isolates of bovine mastitis and the lowest were associated with human infections isolates. There was no relationship between the RAPD patterns and the sources of isolates, except the three clusters which showed host specificity and only included the strains from the same sources. The results confirm the wide genotypic diversity of the studied S. aureus strains. RAPD-PCR technique can be a valuable tool for assessing the genetic relationship, detection of polymorphism in S. aureus and tracing the sources and management of S. aureus infections.
Keywords: Microbiology, Epidemiology, RAPD-PCR, Food, Human, Bovine mastitis, Staphylococcus aureus, Banknotes
1. Introduction
Staphylococcus aureus (S. aureus), a facultative anaerobic gram-positive bacteria, is one of the most significant opportunistic pathogens. In humans, S. aureus is a major cause of foodborne-acquired and hospital-acquired infections. In animals, it elicits mastitis which contributes to major economic loss to the dairy industry (Radwan et al., 2015). It can access milk through direct excretion from udders suffering clinical and sub-clinical staphylococcal mastitis, and by environmental contamination during the milk's handling and processing (Scherrer et al., 2004; Jørgensen et al., 2005). S. aureus commonly harbored by about 30–60% of healthy people in the nares and skin of humans and does not always cause disease (Guidi et al., 2018).
S. aureus is involved in a wide variety of infections found in human and animals, an important cause of bovine mastitis that is one of the most cost-intensive disease in the dairy industry (Rodríguez-Calleja et al., 2006; Morandi et al., 2010; Dufour et al., 2012). Owing to zoonotic potential of S. aureus, control of this bacteria is not only of great economic importance in the dairy industry but also a remarkable public health concern (Kümmel et al., 2016). Some strains are responsible for human food poisoning because of the production of enterotoxins in foodstuffs. Staphylococcal enterotoxins (SEs) that cause abdominal pain, nausea, emesis, and diarrhea, are heat-stable and resistant to human gastrointestinal proteases (Dittmann et al., 2017).
Many phenotypic and genotyping methods have been used in polymorphism analysis including biotyping, multilocus sequence typing (MLST), multiocus variable number tandem repeat analysis (MLVA), pulse field gel electrophoresis (PFGE) and PCR based methods such as random amplified polymorphism DNA (RAPD) PCR, restriction fragment length polymorphic DNA (RFLP) PCR (Hennekinne et al., 2003; Bens et al., 2006; Rabello et al.,2007).
The aim of this study was to assess the genotypic polymorphism among S. aureus isolate were recovered from banknotes, foods, human infections and bovine mastitis milk by RAPD-PCR, and also to investigate the amount of genetic relatedness among these various isolates.
2. Materials and method
2.1. Isolation and identification of S. aureus
A total of fifty S. aureus isolates (12 from banknotes, 12 from foods, 13 from human infections and 13 from bovine mastitis milk) were collected. Banknote isolates were randomly isolated from banknotes of banks in Shiraz, Iran and food isolates were kindly provided by Dr. T. Zahraei Salehi, College of Veterinary Medicine, Tehran University, Iran, which were isolated from restaurants in the University of Tehran. Mastitis isolates were isolated from milk samples of cows with mastitis from farms around Shiraz, Iran, and human infection isolates were collected from patients with nosocomial infections in Namazi Hospital, Shiraz, Iran. Consent form was used to get sample from each patient and the experiment was approved by the ethical committee of the School of Veterinary Medicine, Shiraz University, Shiraz, Iran.
These experiments were biochemically characterized as S. aureus by the conventional biochemical tests such as mannitol fermentation, hydrolysis test of gelatin and urea, protease activity on milk agar medium (Merck, Germany), lipase production on egg yolk agar medium (HiMedia, Mumbia) and hydrolysis of esculin following standard methods (Swan, 1954; Cruickshank et al., 1975). Then stored in Luria-Bertani (LB) broth (Merck, Germany) at -70 °C with 25% glycerol until use (Bair-Parker, 1974).
2.2. DNA extraction and RAPD-PCR analysis
DNA extraction was done using the CinnaPure DNA kit (CinnaGen, Iran) as described by the manufacturer. RAPD-PCR reactions were performed with oligonucleotide primers OLP6 (5′-GAGGGAAGAG-3′), OLP11 (5′-ACGATGAGCC-3′) and OLP13 (5′-ACCGCCTGCT-3′) as described by Williams et al. (1990). Amplifications were carried out in a total volume of 25 μl containing 0.75 μl of oligonucleotides primers (3 μM), 0.75 μl dNTPs (200 μM of each deoxynucleoside triphosphate) (CinnaGen, Iran), 1.5 μl of MgCl2 (3.5 mM) (CinnaGen, Iran), 0.2 μl of Taq DNA polymerase (2.5 U) (CinnaGen, Iran), 2.5 μl of 10X PCR buffer (CinnaGen, Iran), 16.5 μl of sterile distilled water. Rather than “3μl” an estimate of the amount of DNA (ng) is needed.
Amplification of DNA fragments was performed in a MJ Mini Thermal Cycler (BIO-RAD, USA) with initial denaturation at 94 °C for 5 min, followed by 40 cycles of denaturation at 93 °C for 1 min, annealing at 37 °C for 90 S and extension at 72 °C for 1 min, with a final extension of 7 min at 72 °C.
Amplified products were then resolved by electrophoresis in 1.5% agarose gel (CinnaGen, Iran) containing 0.5 μg/ml ethidium bromide (CinnaGen, Iran) and visualized under UV light by Gel Doc (UVitec, UK). A 100 bp DNA ladder (CinnaGen, Iran) was used as a DNA fragment size marker in all gels.
2.3. Fingerprint pattern analysis
The RAPD-PCR banding patterns generated with each primer were analyzed using GelCompar II 6.6.11 software program (Applied Maths, Belgium). Cluster analyses were carried out and dendrograms were created using the unweighted pair-group method with averaging (UPGMA). The DNA relatedness (percentage of similarity) was estimated by using the Dice similarity coefficient.
Isolates are designated genetically indistinguishable if their restriction patterns have the same numbers of bands and the corresponding bands are the same apparent size. Profiles that exhibited ≥94% Dice similarity (≤3 band differences) were considered as closely related strains or clones. Similarity of S. aureus isolates with 65% or greater refereed the probability of the same origin (possibly related isolates) (Tenover et al., 1995).
3. Results
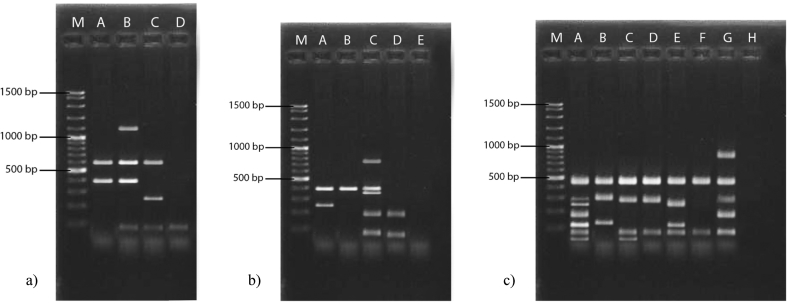
In this study, genotypic polymorphism and the relatedness of fifty S. aureus isolates from banknotes, foods, human infections and bovine mastitis milk were characterized by RAPD-PCR analysis with three different primers (OLP6, OLP11 and OLP13). Amplification of the S. aureus strains with the chosen primers resulted in several polymorphic bands ranging from >50 to >1500 bp in size (Fig. 1).
Fig. 1.
RAPD-PCR patterns of S. aureus isolates from banknotes, foods, human infections and bovine mastitis generated with primers (a) OLP6, (b) OLP11 and (c) OLP13. M: 100 bp DNA ladder.
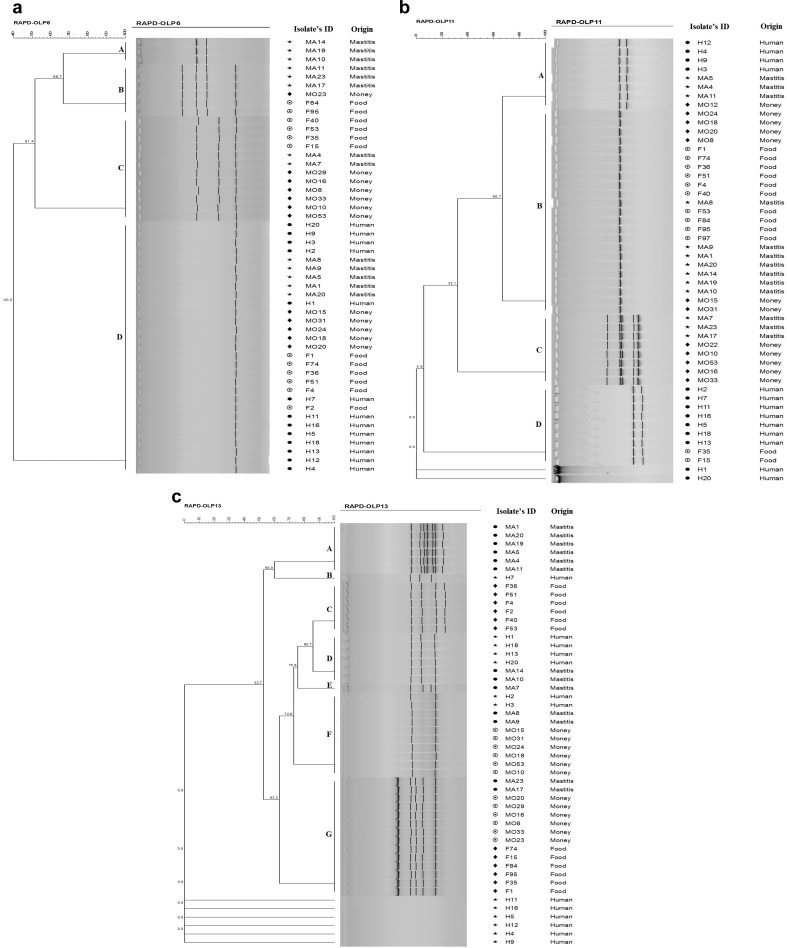
RAPD analysis with primer OLP6 resulted in one to four clear bands (Fig. 1 and Table 1). At 100% RAPD profile similarity value, four major S. aureus clusters (RAPD profiles A-D) including genetically indistinguishable isolates were detected by OLP6 primer (Fig. 2a). Cluster A grouped only three bovine mastitis isolates. Cluster B contained six isolates (three from bovine mastitis, one from banknotes and two from foods), which were possibly related to isolates of cluster A (66.7% RAPD profile similarity). Twelve isolates were distributed in cluster C containing four from foods, two from bovine mastitis and six from banknotes. All isolates from human infections were grouped in Cluster D; this cluster contained 29 isolates (thirteen from human infections, five from bovine mastitis, five from banknotes and six from foods) (Fig. 2a).
Table 1.
Results of RAPD-PCR analysis with OLP6, OLP11 and OLP13 primers on fifty S. aureus isolates.
| Category | Results of RAPD-PCR |
Combination of 3 primers | ||
|---|---|---|---|---|
| OLP6 | OLP11 | OLP13 | ||
| Number of bands | ||||
| Zero | 0 (0%) | 2 (4%) | 6 (12%) | 8/150 (5.33%) |
| One | 29 (58%) | 23 (46%) | 0 (0%) | 52/150 (34.66%) |
| Two | 3 (6%) | 17 (34%) | 10 (20%) | 30/150 (20.0%) |
| Three | 12 (24%) | 0 (0%) | 7 (14%) | 19/150 (12.66%) |
| Four | 6 (12%) | 0 (0%) | 7 (14%) | 13/150 (8.66%) |
| Five | 0 (0%) | 8 (16%) | 14 (28%) | 22/150 (14.66%) |
| Seven | 0 (0%) | 0 (0%) | 6 (12%) | 6/150 (4.0%) |
| Sources of isolates | ||||
| Foods | 3 clusters | 2 clusters | 2 clusters | 7/15 (46.66%) |
| Human infections | 1 cluster | 2 clusters | 3 clusters | 6/15 (40.0%) |
| Bovine mastitis | 4 clusters | 3 clusters | 5 clusters | 12/15 (80.0%) |
| Banknotes | 3 clusters | 3 clusters | 2 clusters | 8/15 (53.33%) |
Fig. 2.
a. Dendrogram of OLP6 RAPD-PCR profiles of fifty S. aureus isolates from banknotes, foods, human infections and bovine mastitis (based on Dice similarity coefficient). b. Dendrogram of OLP11 RAPD-PCR profiles of fifty S. aureus isolates from banknotes, foods, human infections and bovine mastitis (based on Dice similarity coefficient). c. Dendrogram of OLP13 RAPD-PCR profiles of fifty S. aureus isolates from banknotes, foods, human infections and bovine mastitis (based on Dice similarity coefficient).
Amplifications with primer OLP11 yielded zero to five clear bands (Fig. 1 and Table 1). OLP11 oligonucleotide discriminated the fifty S. aureus isolates into four clusters (A-D) of genetically indistinguishable isolates with 100% RAPD profile similarity (Fig. 2b). Cluster A contained 8 isolates (four from human infections, three from bovine mastitis and one from banknotes). Nearly half of S. aureus isolates (23) belonged to cluster B (six from banknotes, ten from foods and seven from bovine mastitis), which can be merged with cluster A to form a cluster containing the majority of S. aureus strains with possibly related genomes (66.7% similarity). Cluster C grouped 8 isolates including three bovine mastitis and five banknote isolates. Cluster D contained 9 isolates (seven from human infections and two from foods). The DNA of two isolates from human infections were not amplified by OLP11 oligonucleotide RAPD-PCR (Fig. 2b).
RAPD analysis with primer OLP13 resulted in two to seven clear bands (Fig. 1 and Table 1). Seven clusters (A-G) containing isolates with 100% RAPD profile similarity could be distinguished by primer OLP13 (Fig. 2c). All the six isolates of cluster A were from bovine mastitis samples. Cluster B only contained one isolate from human infections and cluster C only grouped the six isolates from foods. Four isolates from human infections and two isolates from bovine mastitis belonged to cluster D which had 85.7% similarity with cluster C and 75.6% similarity with cluster E which contained only one isolate from bovine mastitis (possibly related isolates). Cluster F contained 10 isolates (two from human infections, two from bovine mastitis and six from banknotes) which had 72.8% RAPD profile similarity with clusters of C-E. Fourteen isolates were grouped in cluster G (two from bovine mastitis, six from banknotes and six from foods). At 65% similarity cut-off value, the clusters of C–F can be merged together (possibly related isolates). Six isolates from human infections were not amplified by OLP13 RAPD-PCR (Fig. 2c).
4. Discussion
Staphylococcus aureus is a major opportunistic pathogen which is a leading cause of nosocomial infection in human and bovine mastitis and one of the most common agents of food-poisoning (Peton and Le Loir, 2014; Hennekinne et al., 2012; Kadariya et al., 2014; Dittmann et al., 2017). S. aureus has been considered as a great cause of zoonotic disease and the possible transmission amongst livestock and human through close contact, handling and use of infected food of animal origin (Kateete et al., 2013; Wang et al., 2018).
Lack of proper hygienic measures during food preparations such as handling the foods without washing hands leads to contamination of the foods by direct contact or through the respiratory secretions of food handlers who carry pathogenic Staphylococcus. Foods involved with staphylococcal food poisoning are those that are made with hand contact and require no additional cooking (post-processing contamination). These foods are stored at room temperature which allows growth of S. aureus and production of the enterotoxins. S. aureus is also present in food animals and dairy cattle, sheep and goats, and could contaminate the milk of animals with clinical or subclinical mastitis (Argudin et al., 2010).
Banknotes are highly contaminated by pathogenic bacteria and because of its frequent passing from hand to hand, play a role in the transmission of S. aureus to other people through the hand contact. S. aureus are commonly isolated from banknotes from food outlets and hospitals. Simultaneous handling of food and banknotes could lead to the spread of nosocomial infections (Angelakis et al., 2014).
Typing of S. aureus strains may indicate feasible differences in their characteristics also it is helpful to distinguish circulation patterns of these bacteria between several hosts and sources. Many phenotypic and genotypic methods like biotyping, antibiotic susceptibility testing, pulsed-field gel electrophoresis (PFGE), multilocus sequence typing (MLST), and PCR-based techniques including random amplified polymorphisms DNA (RAPD) and restriction fragment length DNA (RFLP) PCR have been used in this way. However, PFGE and MLST are effective methods for typing S. aureus strains, they are expensive, effortful, and technically demanding. Currently, PCR-based typing methods are effective techniques for investigations of strain origin, clonal relatedness among strains and epidemiology. Among these techniques, RAPD-PCR typing is a simple, useful and economically affordable technique which has been widely used for differentiation of S. aureus isolates (Tenover et al., 1994; Van der Zee et al., 1999; Deplano et al., 2000; Neela et al., 2005).
In this study, genetic polymorphism of the S. aureus strains became evident by the RAPD-PCR analysis (Figs. 2a-2c). At 100% similarity level which correspond to genetically indistinguishable isolates, the fifty S. aureus isolates from different sources were classified into four, five and eight groups of the same clone, according to the RAPD-PCR with OLP6, OLP11 and OLP13 primers, respectively.
In the RAPD patterns, some bands were common in all the samples and some were not evident. Maximum amplification was obtained by primer OLP13 which produced seven bands in six bovine mastitis isolates (Table1). Most polymorphisms and strain variability were observed in isolates of bovine mastitis (12 profiles by three primers) and the lowest were associated with human infections isolates (6 profiles by three primers). As in amplification with primer OLP6, all the human infections isolates were clustered together in one group (Fig. 2a).
Dendrograms showed that several isolates could not be distinguished from each other with these oligonucleotides (Figs. 2a-2c). On the basis of which oligonucleotides were used for RAPD-PCR, the strains that clustered together can be different hosts differed. Certain clusters (cluster A in OLP6 and cluster A, C in OLP13 RAPD-PCR) showed host specificity and only included the strains from the same sources. This means that these strains might have emerged from one clone. In addition, cluster B and E in OLP13 RAPD-PCR only included one human infection and one bovine mastitis isolate, respectively. But in other clusters no relationship was found between the RAPD patterns and the sources of isolates.
Result of this study indicates that all the fifty S. aureus isolates were typeable using the combination of the three tested primers in RAPD analysis. Although eight isolates from human infections were untypeable with one primer but were typeable when using another primer (Table 1 and Figs. 2a-2c). These none typeable isolates may have no specific sites in their chromosome for binding of the oligonucleotides primers.
Several studies indicated that S. aureus strains from different hosts were differed from each other and bovine and human isolates were rarely cross-infect (Lange et al., 1999; Larsen et al., 2000; Reinoso et al., 2004). On the other hand, transfer of S. aureus between humans and animals is possible. Milker's hands may have a role in the spread of S. aureus strains associated with bovine mastitis (Reinoso et al., 2004).
In this study, RAPD-PCR technique demonstrated to be efficient in typing and assessment of the genetic relationship of S. aureus isolates from different hosts. The results are in agreement with the earlier report (Reinoso et al., 2004) where, the same primers were successfully applied to assess the genetic relationship between S. aureus isolates from bovine and human hosts. Primers OLP6, OLP11 and OLP13 had high discriminatory power according to the Reinoso et al. (2004) study. In their study, the eighty S. aureus isolates were divided into eleven groups, with most of the human isolates belonging to one group and bovine isolates were distributed over the other ten groups (Reinoso et al., 2004). Fitzgerald et al. (2000) characterized the S. aureus isolates from bovine intramammary infection and reported that the isolates were divided into 12 distinct clonal types on the basis of their RAPD fingerprint profiles.
Several researches have suggested that RAPD-PCR typing is widely used for S. aureus strains (Tambic et al., 1997; Onasanya et al., 2003; Morandi et al., 2010). Some of polymorphisms due to S. aureus are known but those that underscore RAPD-PCR are not so easily identified, and could be because of point mutations in and mobile genetic elements (bacteriophages, plasmids and transposons) (Lindsay, 2010; El-Jakee et al., 2010). In addition, differences in the RAPD-PCR patterns and clustering of S. aureus isolates among the various reports such as Fueyo et al. (2001) and Colombari et al. (2007) studies on S. aureus isolated from human and food samples or Pereira et al. (2002) and Reinoso et al. (2004) studies on human and bovine S. aureus and the current study could be due to use of different oligonucleotide primers, the source of the isolates and occurrence of mutants in S. aureus.
However, the three used primers generated enough polymorphic bands for distinguishing the S. aureus strains, but using the higher number of primers for assessing the genetic relationship between S. aureus isolates had increased the number of different strains.
In conclusion, the results confirm the wide genotypic diversity of S. aureus strains and high dissemination of certain clones through the different host's origins. RAPD-PCR analysis with combination of three primers can be useful for assessing the genetic relationship between S. aureus isolates from different sources. This technique can be a valuable tool for detection of polymorphism in S. aureus and tracing the sources and management of S. aureus infections, and epidemiological studies.
Declarations
Author contribution statement
Sahar Zare: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Abdollah Derakhshandeh, Masoud Haghkhah: Conceived and designed the experiments; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Zahra Naziri, Azar Motamedi Broujeni: Analyzed and interpreted the data.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
The authors would like to thank Dr. Zahra Esmailnezhad for assisting with bacterial isolation.
References
- Angelakis E., Azhar E.I., Bibi F., Yasir M., Al-Ghamdi A.K., Ashshi A.M., Elshemi A.G., Raoult D. Paper money and coins as potential vectors of transmissible disease. Future Microbiol. 2014;9(2):249–261. doi: 10.2217/fmb.13.161. [DOI] [PubMed] [Google Scholar]
- Argudin M.A., Mendoza M.C., Rodicio M.R. Food poisoning and Staphylococcus aureus enterotoxins. Toxins. 2010;2(7):1751–1773. doi: 10.3390/toxins2071751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bair-Parker A.C. Micrococcaceae. In: Buchanan R.E., Gibbons N.E., editors. Bergey’s Manual of Determinative Bacteriology. eighth ed. 1974. pp. 478–490. Baltimore, USA. [Google Scholar]
- Bens C.C.P.M., Voss A., Klaassen C.H.W. Presence of a novel DNA methylation enzyme in methicillin-resistant Staphylococcus aureus isolates associated with pig farming leads to uninterpretable results in standard pulsed-field gel electrophoresis analysis. J. Clin. Microbiol. 2006;44(5):1875–1876. doi: 10.1128/JCM.44.5.1875-1876.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombari V., Mayer M.D., Laicini Z.M., Mamizuka E., Franco B.D., Destro M.T., Landgraf M. Foodborne outbreak caused by Staphylococcus aureus: phenotypic and genotypic characterization of strains of food and human sources. J. Food Prot. 2007;2:279–520. doi: 10.4315/0362-028x-70.2.489. [DOI] [PubMed] [Google Scholar]
- Cruickshank R., Duguid J.P., Marmion B.P., Swain R.H.A. twelfth ed. II. Chruchill Livingstone; Edinburgh, UK: 1975. (Medical Microbiology). [Google Scholar]
- Deplano A., Schuermans A., van Eldere J., Witte W., Meugnier H., Etienne J., Grundmann H., Jonas D., Noordhoek G.T., Dijkstra J., Van Belkum A., Van Leeuwen W., Tassios P.T., Legakis N.J., Van der Zee A., Bergmans A., Blanc D.S., Tenover F.C., Cookson B.C., Oneil G., Struelens M.G. Multicenter evaluation of epidemiological typing of methicillin-resistant Staphylococcus aureus strains by repetitive-element PCR analysis the European Study Group on Epidemiological Markers of the ESCMID. J. Clin. Microbiol. 2000;38(10):3527–3533. doi: 10.1128/jcm.38.10.3527-3533.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmann K.K., Chaul L.T., Lee S.H.I., Corassin C.H., Fernandes de Oliveira C.A., Pereira De Martinis E.C., Alves V.F., Gram L., Oxaran V. Staphylococcus aureus in some Brazilian dairy industries: changes of contamination and diversity. Front. Microbiol. 2017;8:2049. doi: 10.3389/fmicb.2017.02049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufour S., Dohoo I.R., Barkema H.W., Descôteaux L., Devries T.J., Reyher K.K., Roy J.P., Scholl D.T. Manageable risk factors associated with the lactational incidence, elimination, and prevalence of Staphylococcus aureus intramammary infections in dairy cows. J. Dairy Sci. 2012;95:1283–1300. doi: 10.3168/jds.2011-4711. [DOI] [PubMed] [Google Scholar]
- El-Jakee J., Nagwa A.S., Gad El-Said W.A., Bakry M.A., Samy A.A., Khairy E.A., Elgabry E.A. Diversity of Staphylococcus aureus isolated from human and bovine estimated by PCR gene analysis. J. Am. Sci. 2010;6:487–498. [Google Scholar]
- Fitzgerald J.R., Hartigan P.J., Meaney W.J., Smyth C.J. Molecular population and virulence factor analysis of Staphylococcus aureus from bovine intramammary infection. J. Appl. Microbiol. 2000;88:1028–1037. doi: 10.1046/j.1365-2672.2000.01071.x. [DOI] [PubMed] [Google Scholar]
- Fueyo J.M., Martin M.C., Gonzalez-Hevia M.A., Mendoza M.C. Enterotoxin production and DNA fingerprinting in Staphylococcus aureus isolated from human and food samples. Relations between genetic types and enterotoxins. Int. J. Food Microbiol. 2001;67(1–2):139–145. doi: 10.1016/s0168-1605(01)00441-x. [DOI] [PubMed] [Google Scholar]
- Guidi F., Duranti A., Gallina S., Nia Y., Petruzzelli A., Romano A., Travaglini V., Olivastri A., Calvaresi V., Decastelli L., Blasi G. Characterization of A Staphylococcal food poisoning outbreak in A workplace canteen during the post- earthquake reconstruction of Central Italy. Toxins. 2018;10:523. doi: 10.3390/toxins10120523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennekinne J.A., Kerouanton A., Brisabois A., De Buyser M.L. Discrimination of Staphylococcus aureus biotypes by pulsed-field gel electrophoresis of DNA macro-restriction fragments. J. Appl. Microbiol. 2003;94:321–329. doi: 10.1046/j.1365-2672.2003.01837.x. [DOI] [PubMed] [Google Scholar]
- Hennekinne J.A., De Buyser M.L., Dragacci S. Staphylococcus aureus and its food poisoning toxins: characterization and outbreak investigation. FEMS Microbiol. Rev. 2012;36:815–836. doi: 10.1111/j.1574-6976.2011.00311.x. [DOI] [PubMed] [Google Scholar]
- Jørgensen H.J., Mørk T., Rørvik L.M. The occurrence of Staphylococcus aureus on a farm with a small-scale production of raw milk cheese. J. Dairy Sci. 2005;88:3810–3817. doi: 10.3168/jds.S0022-0302(05)73066-6. [DOI] [PubMed] [Google Scholar]
- Kadariya J., Smith T.C., Thapaliya D. Staphylococcus aureus and staphylococcal food-borne disease: an ongoing challenge in public health. BioMed Res. Int. 2014;2014:827965. doi: 10.1155/2014/827965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kateete D.P., Kabugo U., Baluku H., Nyakarahuka L., Kyobe S., Okee M., Nijuaka C.F., Joloba M.L. Prevalence and antimicrobial susceptibility patterns of bacteria from milkmen and cows with clinical mastitis in and around Kampala, Uganda. PLoS One. 2013;8 doi: 10.1371/journal.pone.0063413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kümmel J., Stessl B., Gonano M., Walcher G., Bereuter O., Fricker M., Grunert T., Wagner M., Ehling-Schulz M. Staphylococcus aureus entrance into the dairy chain: tracking S. aureus from dairy cow to cheese. Front. Microbiol. 2016;7:1603. doi: 10.3389/fmicb.2016.01603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange C., Cardoso M., Senczek M., Schwarz S. Molecular subtyping of Staphylococcus aureus isolates from cases of bovine mastitis in Brazil. Vet. Microbiol. 1999;67:127–141. doi: 10.1016/s0378-1135(99)00031-0. [DOI] [PubMed] [Google Scholar]
- Larsen H.D., Huda A., Eriksen N.H., Jensen N.E. Differences between Danish bovine and human Staphylococcus aureus isolates in possession of superantigens. Vet. Microbiol. 2000;76:153–162. doi: 10.1016/s0378-1135(00)00232-7. [DOI] [PubMed] [Google Scholar]
- Lindsay J.A. Genomic variation and evaluation of Staphylococcus aureus. Int. J. Med. Microbiol. 2010;300(2-3):98–103. doi: 10.1016/j.ijmm.2009.08.013. [DOI] [PubMed] [Google Scholar]
- Morandi S., Brasca M., Lodi R., Brusetti L., Andrighetto C., Lombardi A. Biochemical profiles, restriction fragment length polymorphism (RFLP), random amplified polymorphic DNA (RAPD) and multilocus variable number tandem repeat analysis (MLVA) for typing Staphylococcus aureus isolated from dairy products. Res. Vet. Sci. 2010;88:427–435. doi: 10.1016/j.rvsc.2009.10.005. [DOI] [PubMed] [Google Scholar]
- Neela V., Mariana N., Radu S., Zamberi S., Raha A., Rosli R. Use of RAPD to investigate the epidemiology of Staphylococcus aureus infection in Malaysian hospitals. World J. Microbiol. Biotechnol. 2005;21:245–251. [Google Scholar]
- Onasanya A., Mignouna H.D., Thottappilly G. Genetic fingerprinting and phylogenetic diversity of Staphylococcus aureus isolates from Nigeria. Afr. J. Biotechnol. 2003;2(8):246–250. [Google Scholar]
- Pereira M.S.V., Leal N.C., Leal T.C.A., Sobreira M., de Almeida A.M.P., SiqueiraJunior J.P., Campos-Takaki G.M. Typing of human and bovine Staphylococcus aureus by RAPD-PCR and ribotyping-PCR. Lett. Appl. Microbiol. 2002;35:32–36. doi: 10.1046/j.1472-765x.2002.01127.x. [DOI] [PubMed] [Google Scholar]
- Peton V., Le Loir Y. Staphylococcus aureus in veterinary medicine. Infect. Genet. Evol. 2014;21:602–615. doi: 10.1016/j.meegid.2013.08.011. [DOI] [PubMed] [Google Scholar]
- Rabello R.F., Moreira B.M., Lopes R.M., Teixeira L.M., Riley L.W., Castro A.C. Multilocus sequence typing of Staphylococcus aureus isolates recovered from cows with mastitis in Brazilian dairy herds. J. Med. Microbiol. 2007;56:1505–1511. doi: 10.1099/jmm.0.47357-0. [DOI] [PubMed] [Google Scholar]
- Radwan I.A.H., Shehata A.A.E., Abdel-Ghani A.E., Abdullah M.M., Abdraboh A.A.M. Phenotypic and genotypic diversity of Staphylococcus aureus isolated from livestock and human. Glob. Vet. 2015;14(2):274–281. [Google Scholar]
- Reinoso E., Bettera S., Ferigerio C., DiRenazo M., Calozari A., Bongi C. RAPD-PCR analysis of Staphylococcus aureus strains isolated from bovine and human hosts. Microbiol. Res. 2004;159:245–255. doi: 10.1016/j.micres.2004.04.002. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Calleja J.M., García-López I., Santos J.A., Otero A., García-López M.L. Molecular and phenotypic typing of Staphylococcus aureus isolates from rabbit meatRes. An. Microbiol. 2006;157:496–502. doi: 10.1016/j.resmic.2005.11.011. [DOI] [PubMed] [Google Scholar]
- Scherrer D., Corti S., Muehlherr J.E., Zweifel C., Stephan R. Phenotypic and genotypic characteristics of Staphylococcus aureus isolates from raw bulk-tank milk samples of goats and sheep Vet. An. Microbiol. 2004;101:101–107. doi: 10.1016/j.vetmic.2004.03.016. [DOI] [PubMed] [Google Scholar]
- Swan A. The use of a bile-aesculin medium and of Maxted’s technique of Lancefield grouping in the identification of enterococci (Group D Streptococci) J. Clin. Pathol. (Lond.) 1954;7(2):160–163. doi: 10.1136/jcp.7.2.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tambic A., Power E.G., Talsania H., Anthony R.M., French G.L. Analysis of an outbreak of non-phage-typeable methicillin-resistant Staphylococcus aureus by using a randomly amplified polymorphic DNA assay. Vet. Microbiol. 1997;82:61–67. doi: 10.1128/jcm.35.12.3092-3097.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenover F.C., Arbeit R., Archer G., Biddle J., Byrne S., Goering R., Hancock G., Herber G.A., Hill B., Hollis R. Comparison of traditional and molecular methods of typing isolates of Staphylococcus aureus. J. Clin. Microbiol. 1994;32:407–415. doi: 10.1128/jcm.32.2.407-415.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenover F.C., Arbeit R.D., Goering R.V., Mickelsen P.A., Murray B.E., Persing D.H., Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Zee A., Verbakel H., van Zon J.C., Frenay I., van Belkum A., Peeters M., Buiting A., Bergmans A. Molecular genotyping of Staphylococcus aureus strains: comparison of repetitive element sequence-based PCR with various typing methods and isolation of novel epidemicity marker. J. Clin. Microbiol. 1999;37:342. doi: 10.1128/jcm.37.2.342-349.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Lin X., Jiang T., Peng Z., Xu J., Yi L., Li F., Fanning S., Baloch Z. Prevalence and characterization of Staphylococcus aureus cultured from raw milk taken from dairy cows with mastitis in beijing, China. Front. Microbiol. 2018;9:1123. doi: 10.3389/fmicb.2018.01123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams J.G.K., Kubelik A.R., Livak K.J., Rafalski J.A., Tingey S.V. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990;18:6531–6535. doi: 10.1093/nar/18.22.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]