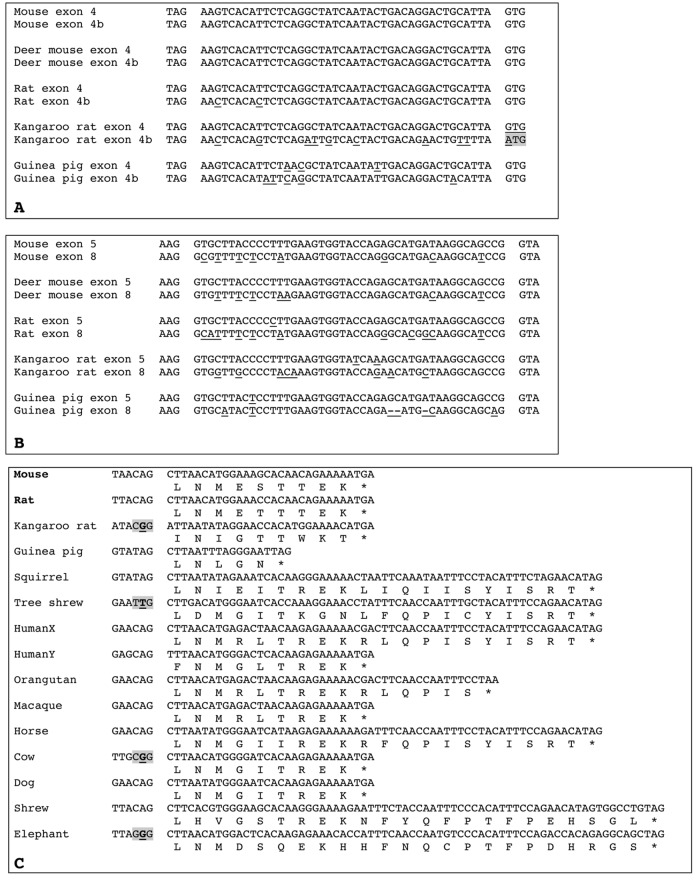
Figure 3.
Alignments of amelogenin exons 4 and 4b, exons 5 and 8, and exon 9. (A) Alignment of AMELX exon 4 with the putative exon “4b” sequence found in 5 rodent genomes. In 4 species, exon “4b” is putatively functional: correct donor (left) and acceptor (right) intron splices and sequence either identical or close to that of exon 4. In the kangaroo rat, exon “4b” is not coding, as indicated by the mutation of the acceptor intron splice (gray background); note that this exon “4b” sequence shows more nucleotide substitutions than in, e.g., mouse and rat sequences. Nucleotide differences between exon 4 and exon “4b” are underlined. Latin names of species are indicated in Appendix 1. (B) Alignment of AMELX exon 5 with exon 8 sequences found in 5 rodent genomes. Mouse and rat sequences are functional. The 3 other exons 8 are putatively functional: correct donor (left) and acceptor (right) intron splices, no deleterious mutations, and sequence close to that of mouse exon 8. Nucleotide differences between exon 5 and exon 8 are underlined. Latin names of species are indicated in Appendix 1. (C) Alignment of functional (i.e., found in mouse and rat AMELX transcripts, in bold), putatively functional (but no cDNA data), or non-coding (i.e., putative intron splice mutated, gray background) AMELX exon 9 (nucleotide and protein sequences) of species representative of various mammalian lineages. Several sequences are putatively functional: correct donor intron splice (shown on the left) and beginning of the sequence similar to that of mouse and rat exon 9. Note the remarkable conservation of the sequence from elephant to mice, and the differences between human AMELX and AMELY sequences. * = stop codon. Latin names of species are indicated in Appendix 1.