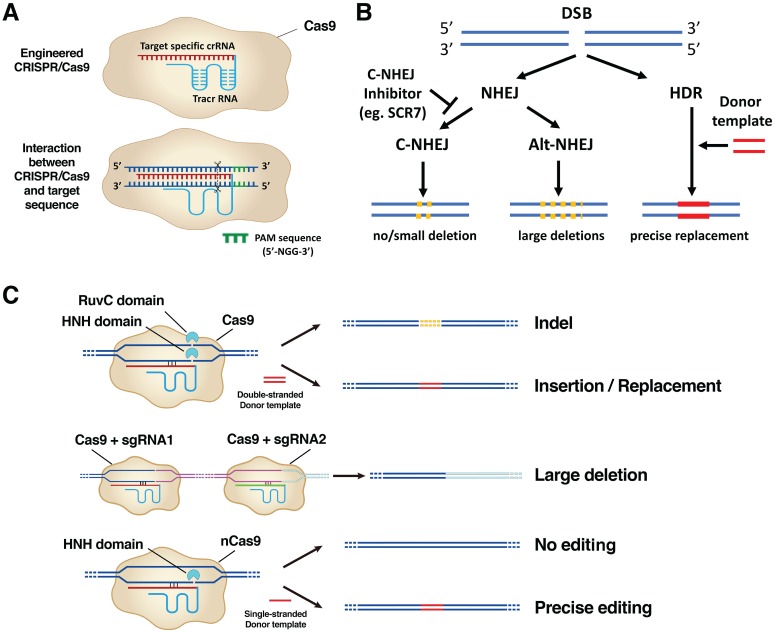
Figure 3.
Mechanisms of the CRISPR/Cas9 system. (A) Top row: Engineered CRISPR/Cas9 is composed of a sgRNA that contains ~20-bp crRNA (red sequence) fused with a tracrRNA (blue sequence) and a Cas9 enzyme component (beige irregular circle). Bottom row: sgRNA binds to the specific target DNA sequences, and Cas9 makes a double-strand break (DSB) at 3 bp upstream of a conserved protospacer-adjacent motif (PAM) region (highlighted in green). Cas9 requires a PAM sequence of 5′-NGG-3′ sequence (N represents any nucleotide base). (B) Repair mechanisms after DSB. Two DNA repair pathways—nonhomologous end-joining (NHEJ) or homology-directed repair (HDR)— kick in after DSB. The NHEJ mechanism is further divided into a canonical (C-NHEJ) or alternative (alt-NHEJ) pathway. C-NHEJ results in no deletion or a small deletion; alt-NHEJ results in the large deletions. Ligase IV inhibitor SCR7 inhibits the C-NHEJ pathway. The HDR pathway is promoted with the presence of a donor template DNA leading to precise replacement of genomic sequences. (C) The application of CRISPR/Cas9 in genome editing. Top row: NHEJ after DSB results in insertion/deletion (indel) mutations; HDR after DSB prompted by a double-stranded donor template DNA results in precise insertion/replacement. Middle row: NHEJ results in a large deletion (pink sequence) when 2 distant DSBs are created by CRISPR/Cas9 systems containing sgRNA1 and sgRNA2. Bottom row: CRISPR/Cas9 with a mutation in the RuvC domain generates nick instead of DSB. DNA repair from a nick with a single-stranded donor template is highly accurate. bp, base pair; crRNA, CRISPR RNA; sgRNA, single-guide RNA; tracrRNA, transactivating crRNA.