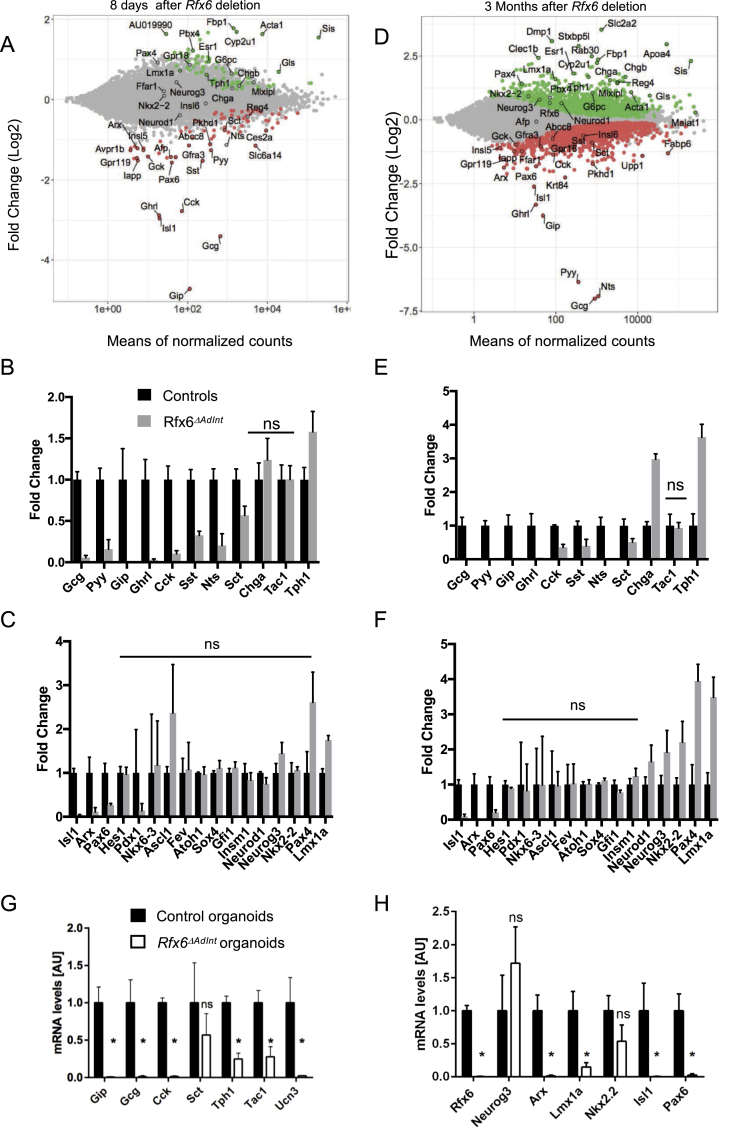
Figure 4.
Gene expression profiling of the small intestine of adult Rfx6ΔAdIntmice. (A–F) RNA-Seq analysis of the small intestine (ileum) of adult Rfx6ΔAdInt mice 8 days (A–C) and 3 months (D–F) after Rfx6 deletion. n = 3–4 animals were analyzed per group. (A, D) MA-plot representing the estimated log2 Fold-Change as a function of the mean of normalized counts. Green and red points represent significantly (adjusted p-value ≤ 0.05) up- and down-regulated genes, respectively in Rfx6ΔAdInt versus controls, 8 days (A) or 3 months (D) after Rfx6 deletion (RNA-Seq data). (B, C, E, F) Histograms showing RNA-Seq data for a selection of genes encoding enteroendocrine hormones or the rate limiting enzyme Tph1 for serotonin synthesis (B, E) or transcription factors (C, F). All controls values are adjusted to 1. ns: non-significant, adjusted p-value >0.05 between controls and mutant. For all other genes adjusted p-value is ≤ 0.05. (G, H) RT-qPCR expression analysis of relevant genes in Rfx6ΔAdint duodenal organoids, 8 days after a 15 h treatment with 1.25 μM tamoxifen (4-OHT) to delete Rfx6 gene. Data are represented as mean ± SD. Organoids were generated from n = 4 mice per group. Statistics: non-parametric Mann–Whitney test, *p ≤ 0.05, ns, non significant.